3WFA
 
 | | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase | | Descriptor: | Alpha-glucosidase, SODIUM ION, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Okuyama, M, Yoshida, T, Hondoh, H, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase
To be Published
|
|
3WEL
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltotriose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3WEN
 
 | | Sugar beet alpha-glucosidase with acarviosyl-maltopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, ... | | Authors: | Tagami, T, Yamashita, K, Okuyama, M, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-09 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural advantage of sugar beet alpha-glucosidase to stabilize the Michaelis complex with long-chain substrate
J.Biol.Chem., 290, 2014
|
|
3VJF
 
 | | Crystal structure of de novo 4-helix bundle protein WA20 | | Descriptor: | POTASSIUM ION, WA20 | | Authors: | Arai, R, Kimura, A, Kobayashi, N, Matsuo, K, Sato, T, Wang, A.F, Platt, J.M, Bradley, L.H, Hecht, M.H. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20
J.Phys.Chem.B, 116, 2012
|
|
2ZID
 
 | | Crystal structure of dextran glucosidase E236Q complex with isomaltotriose | | Descriptor: | CALCIUM ION, Dextran glucosidase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
2ZIC
 
 | | Crystal structure of Streptococcus mutans dextran glucosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Dextran glucosidase, ... | | Authors: | Hondoh, H, Saburi, W, Mori, H, Okuyama, M, Nakada, T, Matsuura, Y, Kimura, A. | | Deposit date: | 2008-02-14 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of alpha-1,6-glucosidic linkage hydrolyzing enzyme, dextran glucosidase from Streptococcus mutans.
J.Mol.Biol., 378, 2008
|
|
3GWF
 
 | | Open crystal structure of cyclohexanone monooxygenase | | Descriptor: | Cyclohexanone monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mirza, I.A, Yachnin, B.J, Berghuis, A.M. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor
J.Am.Chem.Soc., 131, 2009
|
|
3GWD
 
 | | Closed crystal structure of cyclohexanone monooxygenase | | Descriptor: | Cyclohexanone monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mirza, I.A, Yachnin, B.J, Berghuis, A.M. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of cyclohexanone monooxygenase reveal complex domain movements and a sliding cofactor
J.Am.Chem.Soc., 131, 2009
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
3CW4
 
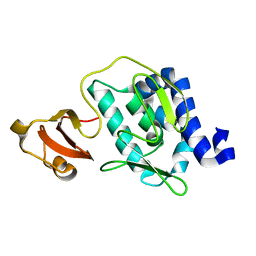 | | Large c-terminal domain of influenza a virus RNA-dependent polymerase PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Kuzuhara, T, Kise, D, Yoshida, H, Horita, T, Murasaki, Y, Utsunomiya, H, Fujiki, H, Tsuge, H. | | Deposit date: | 2008-04-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue
J.Biol.Chem., 284, 2009
|
|
4WLC
 
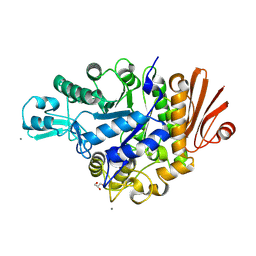 | | Structure of dextran glucosidase with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,6-alpha-glucosidase, ... | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
6KOS
 
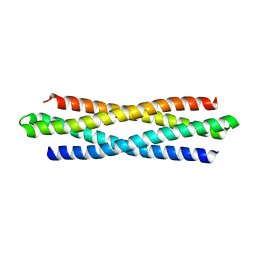 | |
1GLV
 
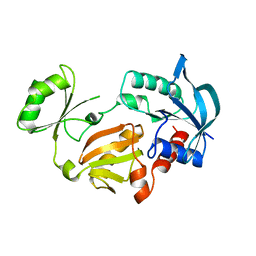 | |
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
5X7G
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, GLYCEROL, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
5X7H
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, MALONATE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
5B3N
 
 | |
1GSA
 
 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
3KGL
 
 | | Crystal structure of procruciferin, 11S globulin from Brassica napus | | Descriptor: | Cruciferin, GLYCEROL, SULFATE ION | | Authors: | Tandang-Silvas, M.R, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
4XB3
 
 | | Structure of dextran glucosidase | | Descriptor: | CALCIUM ION, Glucan 1,6-alpha-glucosidase, HEXAETHYLENE GLYCOL | | Authors: | Kobayashi, M, Kato, K, Yao, M. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structural insights into the catalytic reaction that is involved in the reorientation of Trp238 at the substrate-binding site in GH13 dextran glucosidase
Febs Lett., 589, 2015
|
|
8WX3
 
 | |
8WX1
 
 | |
8J5L
 
 | |
8J5M
 
 | |
8J3M
 
 | |
