7ATD
 
 | | Structure of inactive EstD11 S144A in complex with methyl-naproxen | | Descriptor: | ACETATE ION, EstD11 S144A, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AUY
 
 | |
7ATQ
 
 | | Structure of EstD11 in complex with cyclohexane carboxylic acid | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT2
 
 | |
2IXV
 
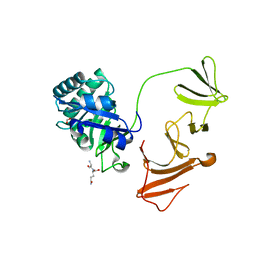 | |
7AJZ
 
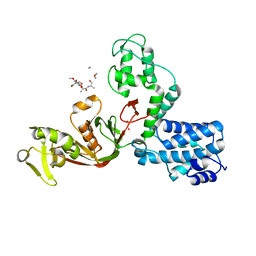 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide) | | Descriptor: | 1,2-ETHANEDIOL, L,D-transpeptidase YcbB, NAG-NAM(tetrapeptide), ... | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide)
To Be Published
|
|
7AGZ
 
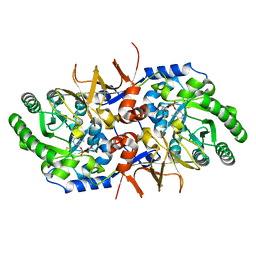 | | BsrV no-histagged | | Descriptor: | Broad specificity amino-acid racemase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Espaillat, A, Cava, F, Hermoso, J.A. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Binding of non-canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity.
Comput Struct Biotechnol J, 19, 2021
|
|
7AJO
 
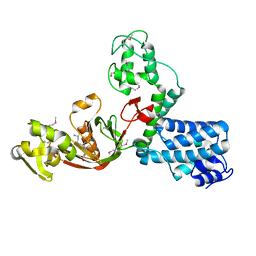 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate | | Descriptor: | (2~{S},6~{S})-2-azanyl-6-[[(4~{R})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]heptanedioic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate
To Be Published
|
|
7AJX
 
 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem
To Be Published
|
|
7AJ9
 
 | |
7AZR
 
 | |
7AZQ
 
 | |
2J8G
 
 | |
2IXU
 
 | |
2J8F
 
 | |
2YP6
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
3ESX
 
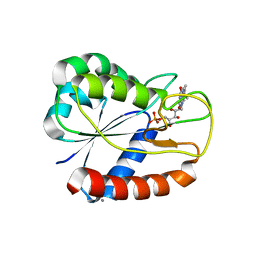 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
8A39
 
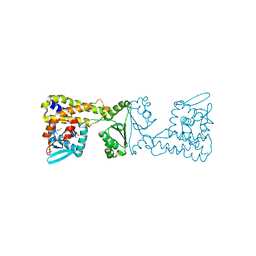 | | Crystal Structure of PaaX from Escherichia coli W | | Descriptor: | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive, GLYCEROL, ... | | Authors: | Molina, R, Alba-Perez, A, Hermoso, J.A. | | Deposit date: | 2022-06-07 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of PaaX, the main repressor of the phenylacetate degradation pathway in Escherichia coli W: A novel fold of transcription regulator proteins.
Int.J.Biol.Macromol., 254, 2024
|
|
8B58
 
 | |
8BRE
 
 | |
2BGI
 
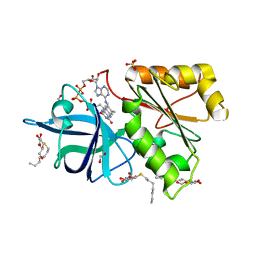 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus complexed with three molecules of the detergent n-heptyl- beta-D-thioglucoside at 1.7 Angstroms | | Descriptor: | CARBON DIOXIDE, FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Ferredoxin-Nadp(H) Reductase from Rhodobacter Capsulatus: Molecular Structure and Catalytic Mechanism
Biochemistry, 44, 2005
|
|
7O4A
 
 | |
7O49
 
 | |
7O4C
 
 | |
