8BPR
 
 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | Descriptor: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2W84
 
 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
2XRY
 
 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
4CDN
 
 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
8A8J
 
 | | Complex of RecF and DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A93
 
 | | Complex of RecF-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3SNL
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
1SKV
 
 | | Crystal Structure of D-63 from Sulfolobus Spindle Virus 1 | | Descriptor: | Hypothetical 7.5 kDa protein | | Authors: | Kraft, P, Kummel, D, Oeckinghaus, A, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of d-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function
J.Virol., 78, 2004
|
|
8AB0
 
 | | Complex of RecO-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA repair protein RecO, Oligo1, Oligo2, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1TBX
 
 | | Crystal structure of SSV1 F-93 | | Descriptor: | Hypothetical 11.0 kDa protein | | Authors: | Kraft, P, Oeckinghaus, A, Kummel, D, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-05-20 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein.
J.Virol., 78, 2004
|
|
3SNI
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3MNQ
 
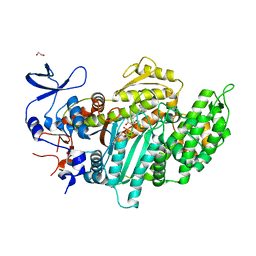 | | Crystal structure of myosin-2 motor domain in complex with ADP-metavanadate and resveratrol | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Schneider, J, Taft, M, Backhaus, A, Baruch, P, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of resveratrol regulation of myosin activity.
To be Published
|
|
6FFC
 
 | | Structure of an inhibitor-bound ABC transporter | | Descriptor: | ATP-binding cassette sub-family G member 2, ~{tert}-butyl 3-[(2~{S},5~{S},8~{S})-14-cyclopentyloxy-2-(2-methylpropyl)-4,7-bis(oxidanylidene)-3,6,17-triazatetracyclo[8.7.0.0^{3,8}.0^{11,16}]heptadeca-1(10),11,13,15-tetraen-5-yl]propanoate | | Authors: | Jackson, S.M, Manolaridis, I, Kowal, J, Zechner, M, Taylor, N.M.I, Bause, M, Bauer, S, Bartholomaeus, R, Stahlberg, H, Bernhardt, G, Koenig, B, Buschauer, A, Altmann, K.H, Locher, K.P. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of small-molecule inhibition of human multidrug transporter ABCG2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2J08
 
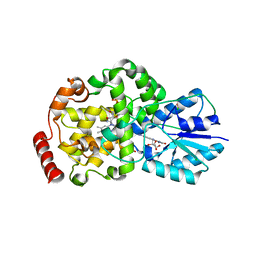 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J09
 
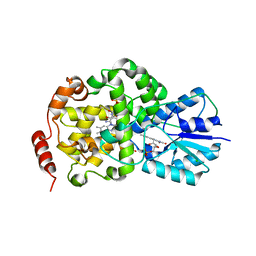 | | Thermus DNA photolyase with FMN antenna chromophore | | Descriptor: | CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J07
 
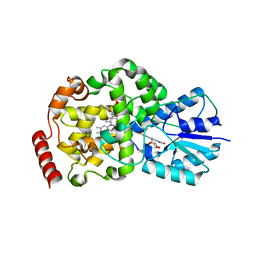 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2K9C
 
 | | Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Balayssac, S, Bertini, I, Bhaumik, A, Lelli, M, Luchinat, C. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic shifts in solid-state NMR of proteins to elicit structural information
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KRJ
 
 | | High-Resolution Solid-State NMR Structure of a 17.6 kDa Protein | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Bertini, I, Bhaumik, A, De Pa pe, G, Griffin, R.G, Lelli, M, Lewandowski, J.R, Luchinat, C. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution solid-state NMR structure of a 17.6 kDa protein.
J.Am.Chem.Soc., 132, 2010
|
|
4AN7
 
 | |
4BXU
 
 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
8G8Z
 
 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | Authors: | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5G1V
 
 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1W
 
 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
