6KUP
 
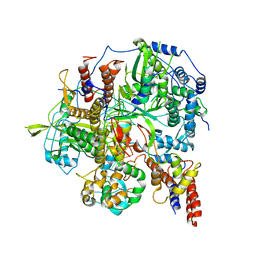 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
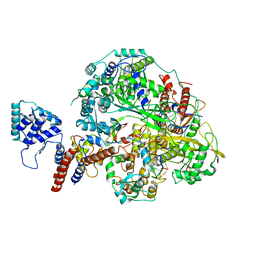 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
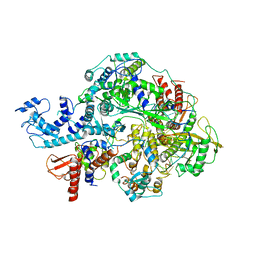 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
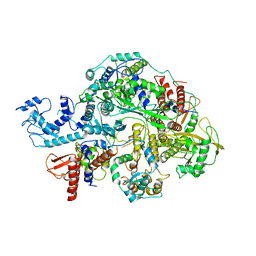 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
5XN4
 
 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
6KUJ
 
 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 1 | | Descriptor: | 3'-cRNA promoter, 5'-cRNA promoter, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of influenza D virus polymerase bound to cRNA promoter in Mode A conformation
NAT NANOTECHNOL, 2019
|
|
7KHD
 
 | | Human GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.956102 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7KHX
 
 | | Mouse GITR-GITRL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, ... | | Authors: | Wang, F, Chau, B, West, S.M, Strop, P. | | Deposit date: | 2020-10-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.20569849 Å) | | Cite: | Structures of mouse and human GITR-GITRL complexes reveal unique TNF superfamily interactions.
Nat Commun, 12, 2021
|
|
7M7D
 
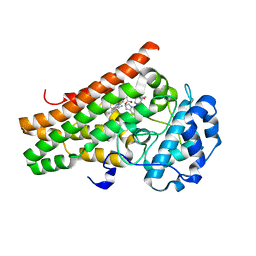 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
7M63
 
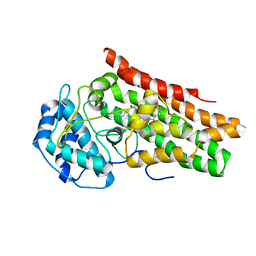 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-70099 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[(1R,3S,5S,6r)-3-(5,6-difluoro-1H-benzimidazol-1-yl)bicyclo[3.1.0]hexan-6-yl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
6S1F
 
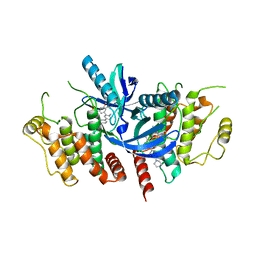 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
7OWD
 
 | |
7OWC
 
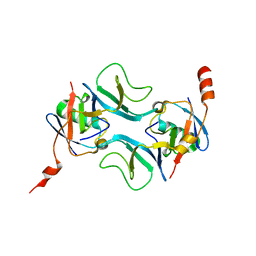 | |
6IS2
 
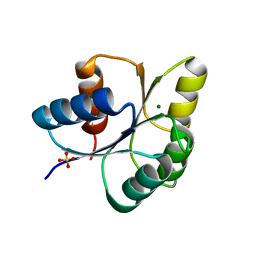 | |
6IS3
 
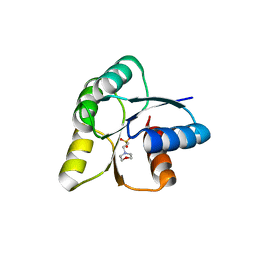 | |
5YQZ
 
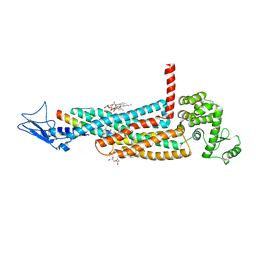 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
6IS4
 
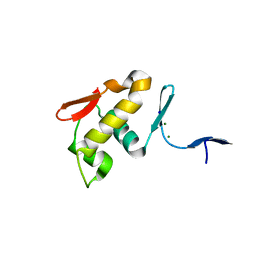 | |
6IS1
 
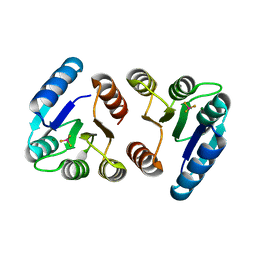 | |
6FU5
 
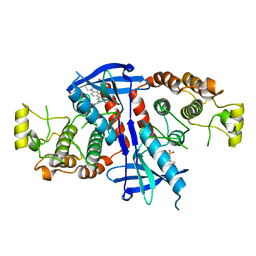 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP18 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[5-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-2-methoxy-phenyl]propane-1-sulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling.
EMBO J., 37, 2018
|
|
4C8B
 
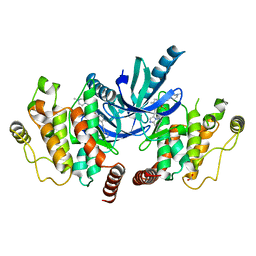 | | Structure of the kinase domain of human RIPK2 in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Canning, P, Krojer, T, Bradley, A, Mahajan, P, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors.
Chem. Biol., 22, 2015
|
|
6L5K
 
 | | ARF5 Aux/IAA17 Complex | | Descriptor: | Auxin response factor 5, Auxin-responsive protein IAA17 | | Authors: | Ryu, K.S, Suh, J.Y, Cha, S.Y, Kim, Y.I, Park, C.K. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Determinants of PB1 Domain Interactions in Auxin Response Factor ARF5 and Repressor IAA17.
J.Mol.Biol., 432, 2020
|
|
2Q2O
 
 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q3J
 
 | | Crystal structure of the His183Ala variant of Bacillus subtilis ferrochelatase in complex with N-Methyl Mesoporphyrin | | Descriptor: | Ferrochelatase, MAGNESIUM ION, N-METHYL PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-30 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q2N
 
 | | Crystal structure of Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | Descriptor: | Non-structural protein 1 | | Authors: | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
