6M8C
 
 | |
6M8A
 
 | |
6M8E
 
 | |
6MXY
 
 | |
6MXZ
 
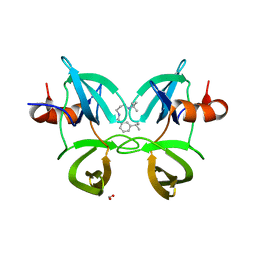 | | Structure of 53BP1 Tudor domains in complex with small molecule UNC3474 | | Descriptor: | FORMIC ACID, N-[3-(tert-butylamino)propyl]-3-(propan-2-yl)benzamide, TP53-binding protein 1 | | Authors: | Cui, G, Botuyan, M.V, Schuller, D.J, Mer, G. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering.
Nat Commun, 14, 2023
|
|
5CZK
 
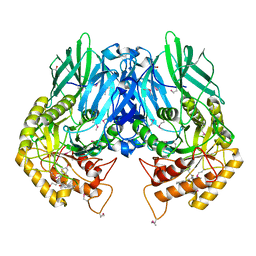 | | Structure of E. coli beta-glucuronidase bound with a novel, potent inhibitor 1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea | | Descriptor: | 1-[(6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Roberts, A.R, Wallace, B.R, Redinbo, M.R. | | Deposit date: | 2015-07-31 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity.
Chem.Biol., 22, 2015
|
|
6V2R
 
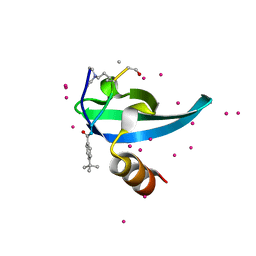 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
