4B7A
 
 | | Probing the active center of catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, CALCIUM ION, CATALASE-PHENOL OXIDASE, ... | | Authors: | Yuzugullu, Y, Trinh, C.H, Pearson, A.R, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the Active Center of Catalase-Phenol Oxidase from Scytalidium Thermophilum
To be Published
|
|
4BCU
 
 | | Satellite Tobacco Necrosis Virus (STNV) virus like particle in complex with the B3 aptamer | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Ford, R.J, Barker, A.M, Bakker, S.E, Coutts, R.H, Ranson, N.A, Phillips, S.E.V, Pearson, A.R, Stockley, P.G. | | Deposit date: | 2012-10-03 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Sequence-Specific, RNA-Protein Interactions Overcome Electrostatic Barriers Preventing Assembly of Satellite Tobacco Necrosis Virus Coat Protein.
J.Mol.Biol., 425, 2013
|
|
4C3C
 
 | | Thaumatin refined against hatrx data for time-point 1 | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN-1 | | Authors: | Yorke, B.A, Beddard, G.S, Owen, R.L, Pearson, A.R. | | Deposit date: | 2013-08-22 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Resolved Crystallography Using the Hadamard Transform.
Nat.Methods, 11, 2014
|
|
4CRZ
 
 | | Direct visualisation of strain-induced protein prost-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4CIZ
 
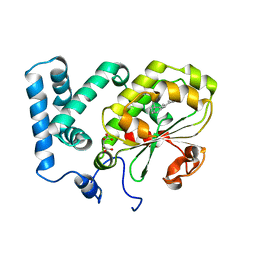 | | Crystal structure of the complex of the Cellular Retinal Binding Protein with 9-cis-retinal | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4CJ6
 
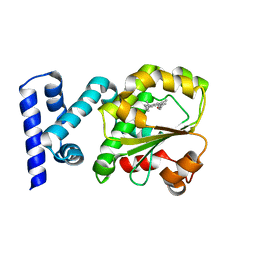 | | Crystal structure of the complex of the Cellular Retinal Binding Protein Mutant R234W with 9-cis-retinal | | Descriptor: | RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4CS0
 
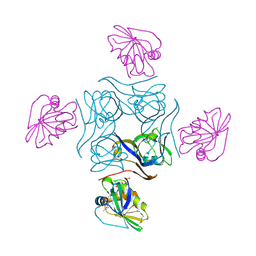 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4AUN
 
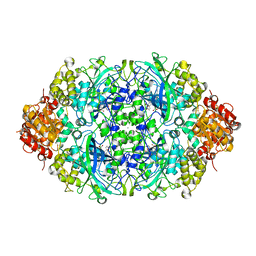 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AZD
 
 | | T57V mutant of aspartate decarboxylase | | Descriptor: | ASPARTATE 1-DECARBOXYLASE, MALONATE ION | | Authors: | Webb, M.E, Yorke, B.A, Kershaw, T, Lovelock, S, Lobley, C.M.C, Kilkenny, M.L, Smith, A.G, Blundell, T.L, Pearson, A.R, Abell, C. | | Deposit date: | 2012-06-25 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Threonine 57 is Required for the Post-Translational Activation of Escherichia Coli Aspartate Alpha-Decarboxylase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4B2Y
 
 | | Probing the active center of catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuzugullu, Y, Trinh, C.H, Pearson, A.R, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-07-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating the Active Centre of the Scytalidium Thermophilum Catalase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4CTF
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | EQUINE RHINITIS A VIRUS, P1, VP1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
4AUL
 
 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4D7Z
 
 | | E. coli L-aspartate-alpha-decarboxylase mutant N72Q to a resolution of 1.9 Angstroms | | Descriptor: | ASPARTATE 1-DECARBOXYLASE ALPHA CHAIN, ASPARTATE 1-DECARBOXYLASE BETA CHAIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bravo, J.P.K, Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2014-12-02 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the E. Coli L-Aspartate-Alpha-Decarboxylase Mutant N72Q to a Resolution of 1.9 Angstroms
To be Published
|
|
4AUM
 
 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
4CTG
 
 | | The limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle | | Descriptor: | P1 | | Authors: | Bakker, S.E, Groppelli, E, Pearson, A.R, Stockley, P.G, Rowlands, D.J, Ranson, N.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Limits of Structural Plasticity in a Picornavirus Capsid Revealed by a Massively Expanded Equine Rhinitis a Virus Particle.
J.Virol., 88, 2014
|
|
4CRY
 
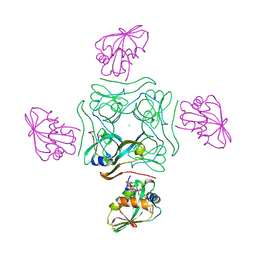 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, CHLORIDE ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Direct Visualisation of Strain-Induced Protein Post-Translational Modification
To be Published
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ZB7
 
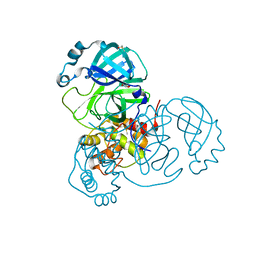 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant Y54F at 1.63 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
7ZB6
 
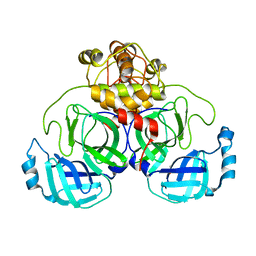 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant C44S at 2.12 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
7ZB8
 
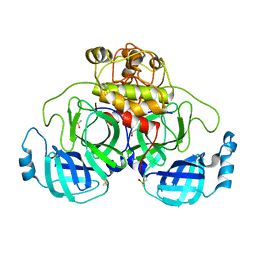 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant K61A at 2.48 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8BD0
 
 | |
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
