6IYM
 
 | |
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
3QID
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
8G36
 
 | | Crystal structure of F182L-CYP199A4 in complex with terephthalic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
8G35
 
 | | Crystal structure of F182L-CYP199A4 in complex with (S)-4-(2-hydroxy-3-oxobutan-2-yl)benzoic acid | | Descriptor: | 4-[(2S)-2-hydroxy-3-oxobutan-2-yl]benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
7JW5
 
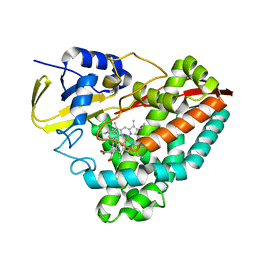 | | Crystal structure of WT-CYP199A4 in complex with 4-phenylbenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Different Geometric Requirements for Cytochrome P450-Catalyzed Aliphatic Versus Aromatic Hydroxylation Results in Chemoselective Oxidation
Acs Catalysis, 12, 2022
|
|
4ZTT
 
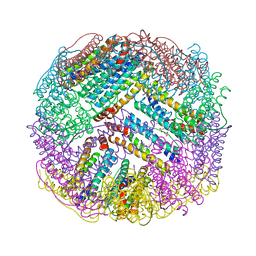 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
4ZZ7
 
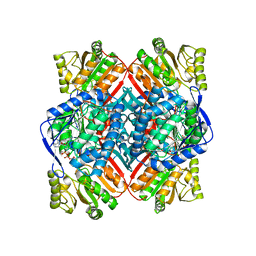 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
8ILJ
 
 | |
6LVP
 
 | |
7DLS
 
 | | Cytochrome P450 (CYP105D18) complex with papaverine | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Characterization of high-H 2 O 2 -tolerant bacterial cytochrome P450 CYP105D18: insights into papaverine N-oxidation.
Iucrj, 8, 2021
|
|
7DI3
 
 | | Cytochrome P450 (CYP105D18) W.T. | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2020-11-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Characterization of high-H 2 O 2 -tolerant bacterial cytochrome P450 CYP105D18: insights into papaverine N-oxidation.
Iucrj, 8, 2021
|
|
8HGU
 
 | |
8HM5
 
 | |
6LVO
 
 | |
8HG9
 
 | | Cytochrome P450 steroid hydroxylase (BaCYP106A6) from Bacillus species | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 steroid hydroxylase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 Steroid Hydroxylase ( Ba CYP106A6) from Bacillus Species.
J Microbiol Biotechnol., 33, 2023
|
|
5X3F
 
 | | Crystal structure of the YgjG-Protein A-Zpa963-PKA catalytic domain | | Descriptor: | Putrescine aminotransferase,Immunoglobulin G-binding protein A, Zpa963,cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2017-02-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5XBY
 
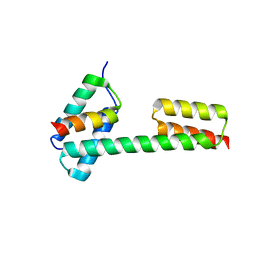 | | Crystal structure of the PKA-Protein A fusion protein (end-to-end fusion) | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2017-03-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
8GTL
 
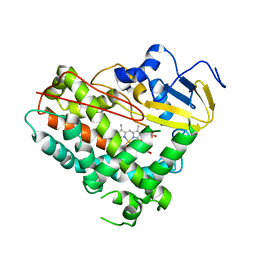 | | Crystal Structure of Cytochrome P450 (CYP101D5) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 CYP101D5 | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 CYP101D5 from Sphingomonas echinoides.
Int J Mol Sci, 23, 2022
|
|
8HEA
 
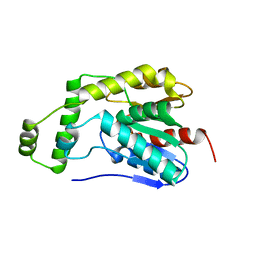 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
5Y7W
 
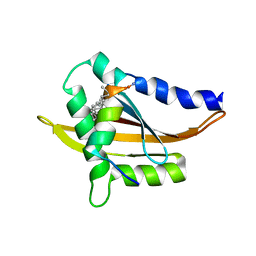 | | Crystal structure of the Nco-A1 PAS-B domain with YL-2 | | Descriptor: | Nuclear receptor coactivator 1, YL-2 peptide | | Authors: | Lee, Y.J, Yoon, H.S, Lee, J.H, Bae, J.H, Song, J.Y, Lim, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Inhibition of the NCOA1/STAT6 Protein-Protein Interaction
J. Am. Chem. Soc., 139, 2017
|
|
8JUO
 
 | |
8JUS
 
 | |
8GJE
 
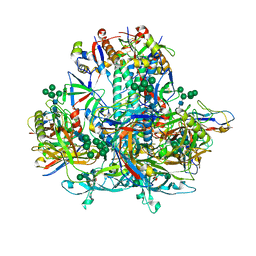 | | HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, J.H, Ward, A.B. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Plos Pathog., 19, 2023
|
|
