5YOC
 
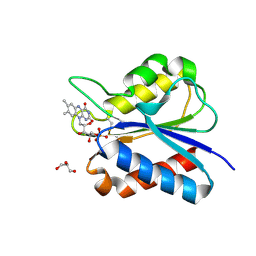 | | Crystal Structure of flavodoxin with engineered disulfide bond C102-R125C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YOG
 
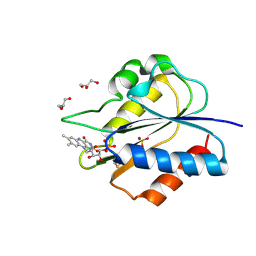 | | Crystal Structure of flavodoxin with engineered disulfide bond N14C-C93 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL, ... | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
6AKU
 
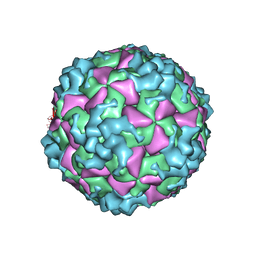 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
2BI8
 
 | |
5YO4
 
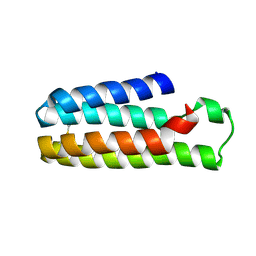 | |
2BI7
 
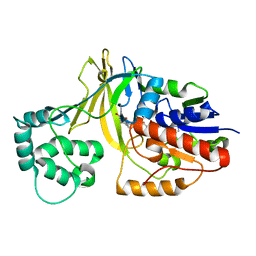 | | udp-galactopyranose mutase from Klebsiella pneumoniae oxidised FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Srikannathasan, V, Naismith, J. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mycobacteria Tuberculosis and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the Oxidised State and Klebsiella Pneumoniae Udp-Galactopyranose Mutase in the (Active) Reduced State.
J.Mol.Biol., 348, 2005
|
|
2HH5
 
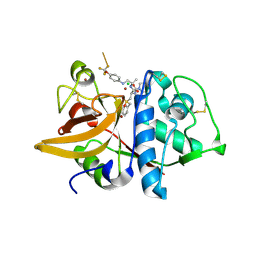 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5YO5
 
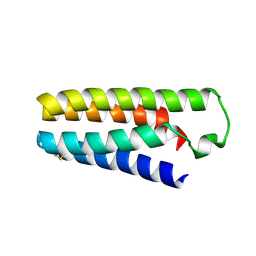 | |
5YOE
 
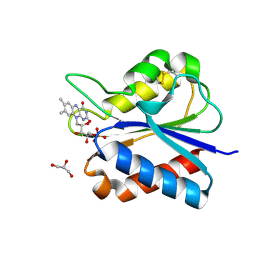 | | Crystal Structure of flavodoxin with engineered disulfide bond A43C-L74C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
2CKG
 
 | |
5YM7
 
 | |
5YO6
 
 | |
5YO3
 
 | | Crystal Structure of B562RIL with engineered disulfide bond V16C-A29C | | Descriptor: | SULFATE ION, Soluble cytochrome b562 | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YOB
 
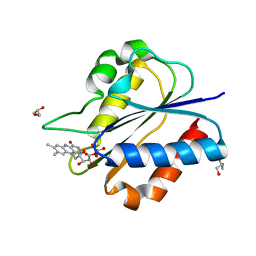 | | Crystal Structure of flavodoxin without engineered disulfide bond | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.142 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
1V0J
 
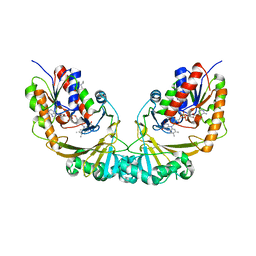 | | Udp-galactopyranose mutase from Mycobacterium tuberculosis | | Descriptor: | BICINE, FLAVIN-ADENINE DINUCLEOTIDE, UDP-GALACTOPYRANOSE MUTASE | | Authors: | Beis, K, Naismith, J.H. | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidised state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state.
J. Mol. Biol., 348, 2005
|
|
2F1G
 
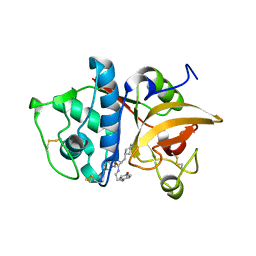 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5HO4
 
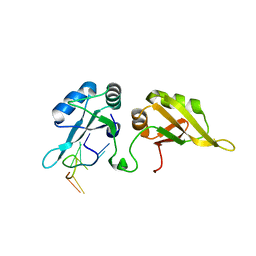 | | Crystal structure of hnRNPA2B1 in complex with 10-mer RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*AP*GP*C)-3') | | Authors: | Wu, B.X, Su, S.C, Gan, J.H, Ma, J.B. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
1BLS
 
 | |
5ID6
 
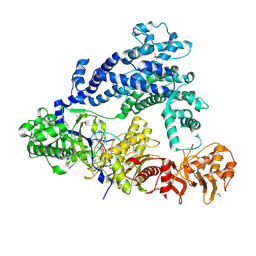 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
5JRE
 
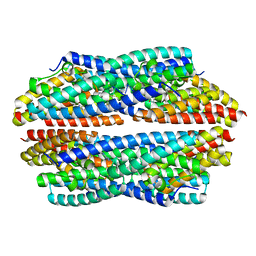 | | Crystal structure of NeC3PO in complex with ssDNA. | | Descriptor: | 9-METHYL-9H-PURIN-6-AMINE, ADENINE, NEQ131, ... | | Authors: | Gan, J, Zhang, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
5XOB
 
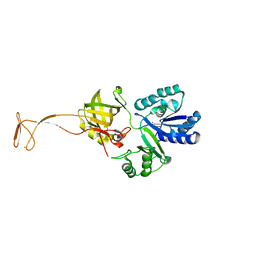 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
5JRC
 
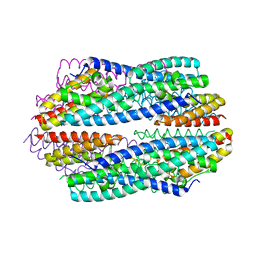 | |
5JQG
 
 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Naismith, J.H, Zhu, X. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
5JR9
 
 | | Crystal structure of apo-NeC3PO | | Descriptor: | NEQ131 | | Authors: | Zhang, J, Gan, J. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for single-stranded RNA recognition and cleavage by C3PO
Nucleic Acids Res., 44, 2016
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
