8X7K
 
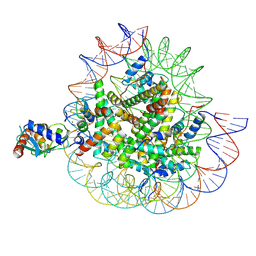 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | Descriptor: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7I
 
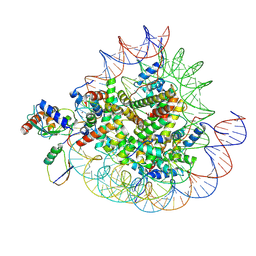 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | Deposit date: | 2023-11-24 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
3FD5
 
 | |
4E93
 
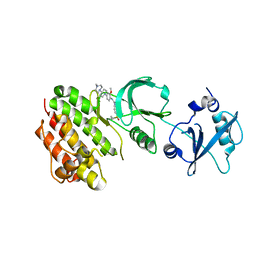 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | Authors: | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
2LB5
 
 | | Refined Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor histidine kinase | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form.
Nature, 463, 2010
|
|
5UDE
 
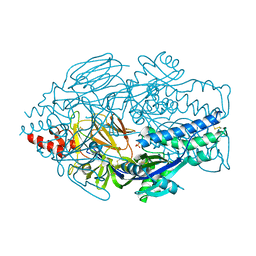 | | Crystal Structure of RSV F B9320 DS-Cav1 | | Descriptor: | Fusion glycoprotein F0, SULFATE ION | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
2LD1
 
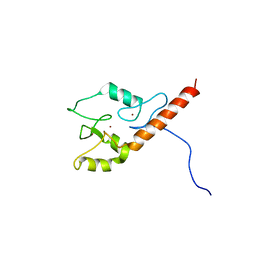 | |
5UDC
 
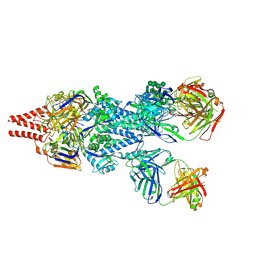 | | Crystal Structure of RSV F A2 Bound to MEDI8897 | | Descriptor: | CHLORIDE ION, Fusion glycoprotein F0, MEDI8897 Fab Heavy Chain, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
2LHR
 
 | |
7F1E
 
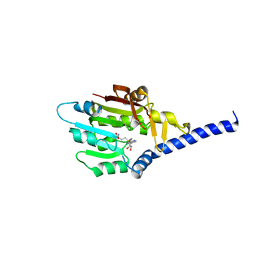 | | Structure of METTL6 bound with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Li, S, Liao, S, Xu, C. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
3L1L
 
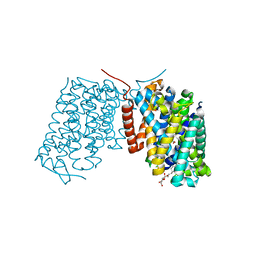 | | Structure of Arg-bound Escherichia coli AdiC | | Descriptor: | ARGININE, Arginine/agmatine antiporter, nonyl beta-D-glucopyranoside | | Authors: | Gao, X, Zhou, L, Shi, Y. | | Deposit date: | 2009-12-13 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Mechanism of substrate recognition and transport by an amino acid antiporter
Nature, 463, 2010
|
|
2BFE
 
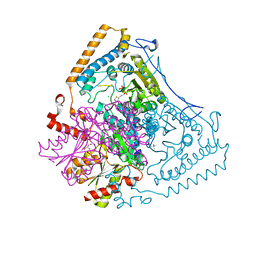 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFD
 
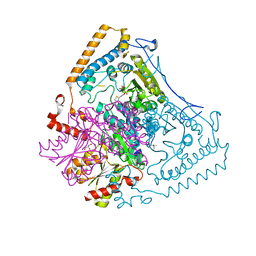 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
6JHY
 
 | |
2LCC
 
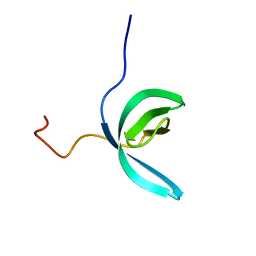 | | Solution structure of RBBP1 chromobarrel domain | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Gong, W, Feng, Y. | | Deposit date: | 2011-04-28 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of methylated histone tails by retinoblastoma-binding protein 1.
J.Biol.Chem., 2012
|
|
2BFC
 
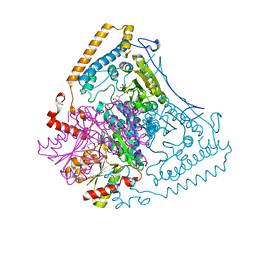 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
4Y16
 
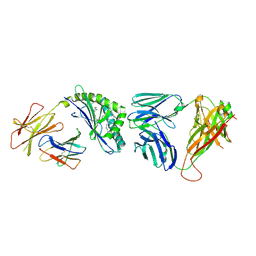 | | Crystal structure of the mCD1d/NC-aGC/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Nemcovic, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lipid and Carbohydrate Modifications of alpha-Galactosylceramide Differently Influence Mouse and Human Type I Natural Killer T Cell Activation.
J.Biol.Chem., 290, 2015
|
|
3L9R
 
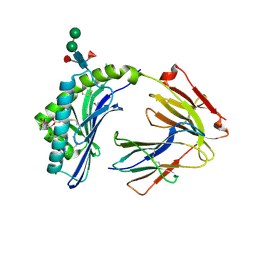 | | Crystal structure of bovine CD1b3 with endogenously bound ligands | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Girardi, E. | | Deposit date: | 2010-01-05 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of bovine CD1b3 with endogenously bound ligands.
J.Immunol., 185, 2010
|
|
4IRE
 
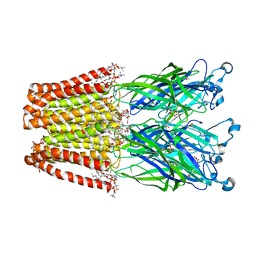 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
8JB0
 
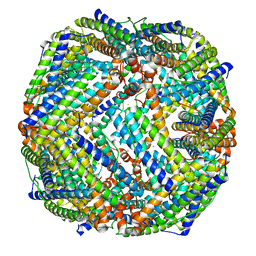 | |
6Z3C
 
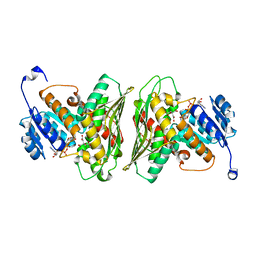 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
8JAX
 
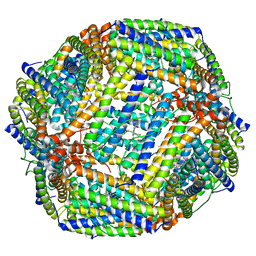 | |
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
6TR3
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 in complex with fucose | | Descriptor: | CALCIUM ION, F5/8 type C domain-containing protein, MAGNESIUM ION, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6TR4
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, F5/8 type C domain-containing protein, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
