4COM
 
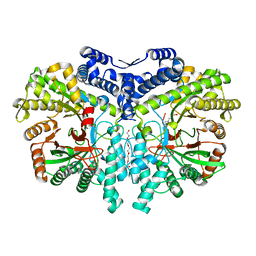 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
1GHE
 
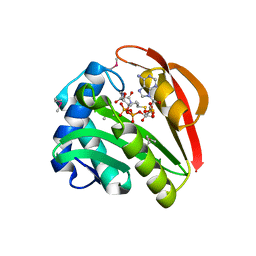 | | CRYSTAL STRUCTURE OF TABTOXIN RESISTANCE PROTEIN COMPLEXED WITH AN ACYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, ACETYLTRANSFERASE | | Authors: | He, H, Ding, Y, Bartlam, M, Sun, F, Le, Y, Qin, X, Tang, H, Zhang, R, Joachimiak, A, Liu, Y, Zhao, N, Rao, Z. | | Deposit date: | 2000-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Tabtoxin Resistance Protein Complexed with Acetyl
Coenzyme A Reveals the Mechanism for beta-Lactam Acetylation
J.Mol.Biol., 325, 2003
|
|
6JGY
 
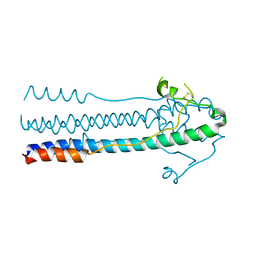 | | Crystal structure of LASV-GP2 in a post fusion conformation | | Descriptor: | Pre-glycoprotein polyprotein GP complex | | Authors: | Zhu, Y, Zhang, X, Chen, B, Ye, S, Zhang, R. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Crystal Structure of Refolding Fusion Core of Lassa Virus GP2 and Design of Lassa Virus Fusion Inhibitors.
Front Microbiol, 10, 2019
|
|
3RQF
 
 | |
2VC0
 
 | |
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
2W25
 
 | | Crystal structure of Glu104Ala mutant | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, RAmachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
2W24
 
 | | M. tuberculosis Rv3291c complexed to Lysine | | Descriptor: | LYSINE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Ramachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
1YMZ
 
 | | CC45, An Artificial WW Domain Designed Using Statistical Coupling Analysis | | Descriptor: | CC45 | | Authors: | Socolich, M, Lockless, S.W, Russ, W.P, Lee, H, Gardner, K.H, Ranganathan, R. | | Deposit date: | 2005-01-22 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolutionary information for specifying a protein fold.
Nature, 437, 2005
|
|
3RGL
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine | | Descriptor: | (2S)-2-hydroxybutanedioic acid, ADENOSINE-5'-TRIPHOSPHATE, GLYCINE, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-08 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP and glycine.
To be Published
|
|
6JKK
 
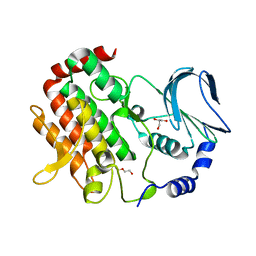 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
3ZS1
 
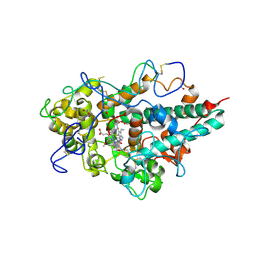 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
6JMU
 
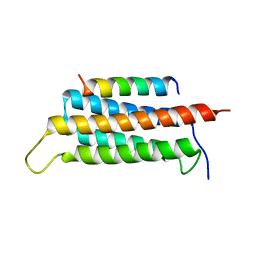 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
2VU5
 
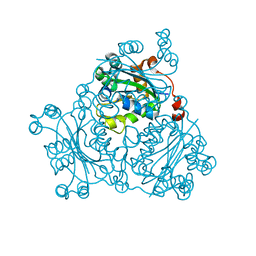 | | Crystal structure of Pndk from Bacillus anthracis | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Misra, G, Aggarwal, A, Dube, D, Zaman, M.S, Singh, Y, Ramachandran, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bacillus Anthracis Nucleoside Diphosphate Kinase and its Characterization Reveals an Enzyme Adapted to Perform Under Stress Conditions.
Proteins, 76, 2009
|
|
3RQG
 
 | |
2VBY
 
 | |
1ZG8
 
 | | Crystal Structure of (R)-2-(3-{[amino(imino)methyl]amino}phenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | (2R)-2-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
4COJ
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima in complex with dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
1FYG
 
 | |
4COI
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with glycerol in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, GLYCEROL, ZINC ION | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
1ZG9
 
 | | Crystal Structure of 5-{[amino(imino)methyl]amino}-2-(sulfanylmethyl)pentanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 5-{[AMINO(IMINO)METHYL]AMINO}-2-(SULFANYLMETHYL)PENTANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
3S81
 
 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
3ZS0
 
 | | Human Myeloperoxidase inactivated by TX2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-FLUOROBENZYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
1ZG7
 
 | | Crystal Structure of 2-(5-{[amino(imino)methyl]amino}-2-chlorophenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 2-(5-{[AMINO(IMINO)METHYL]AMINO}-2-CHLOROPHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
2VC1
 
 | |
