3MJD
 
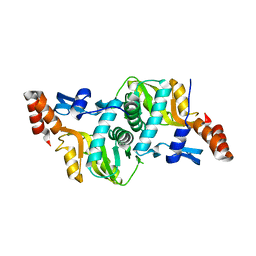 | | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Orotate phosphoribosyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis.
TO BE PUBLISHED
|
|
3M84
 
 | | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from
Francisella tularensis
To be Published
|
|
3MXT
 
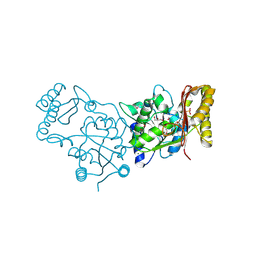 | | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-07 | | Release date: | 2010-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Pantoate-Beta-alanine Ligase from Campylobacter jejuni
To be Published
|
|
3N2B
 
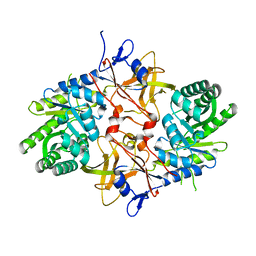 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
3N3W
 
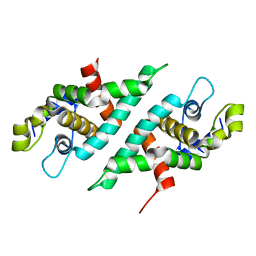 | | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni
TO BE PUBLISHED
|
|
3N08
 
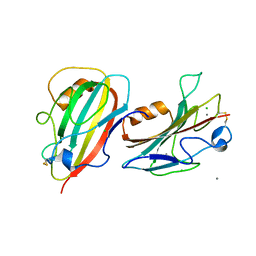 | | Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative PhosphatidylEthanolamine-Binding Protein (PEBP) | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX
To be Published
|
|
3N2L
 
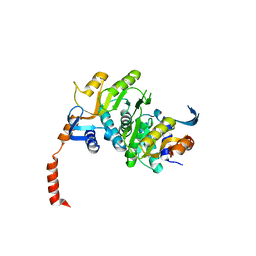 | | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase (pyrE) from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase
(pyrE) from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3MZZ
 
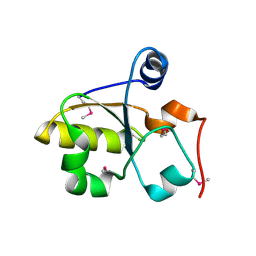 | | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus | | Descriptor: | Rhodanese-like domain protein | | Authors: | Kim, Y, Chruszcz, M, Minor, W, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Rhodanese-like Domain Protein from Staphylococcus aureus
To be Published
|
|
3M5W
 
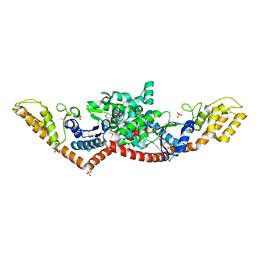 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-14 | | Release date: | 2010-03-31 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni
To be Published, 2010
|
|
3M7W
 
 | | Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 in Covalent Complex with Dehydroquinate | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
3M49
 
 | | Crystal Structure of Transketolase Complexed with Thiamine Diphosphate from Bacillus anthracis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-10 | | Release date: | 2010-04-07 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Transketolase Complexed with Thiamine Diphosphate from Bacillus anthracis
To be Published
|
|
3MGA
 
 | | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Enterochelin esterase, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Ferric Enterobactin Esterase (fes) from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3M5U
 
 | | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Phosphoserine aminotransferase | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni
To be Published
|
|
3MJF
 
 | | Phosphoribosylamine-glycine ligase from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystal structure of phosphoribosylamine-glycine ligase from Yersinia pestis.
To be Published
|
|
3N4J
 
 | | Putative RNA methyltransferase from Yersinia pestis | | Descriptor: | RNA methyltransferase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
3MUE
 
 | | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium | | Descriptor: | ACETIC ACID, ETHANOL, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Pantoate-beta-Alanine Ligase from Salmonella typhimurium
To be Published
|
|
3N4K
 
 | | Putative RNA methyltransferase from Yersinia pestis in complex with S-ADENOSYL-L-HOMOCYSTEINE. | | Descriptor: | RNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Osipiuk, J, Maltseva, N, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray crystal structure of putative RNA methyltransferase from Yersinia pestis.
To be Published
|
|
3N2I
 
 | | 2.25 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with thymidine | | Descriptor: | CHLORIDE ION, THYMIDINE, Thymidylate kinase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with thymidine
To be Published
|
|
4HS7
 
 | | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG. | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bacterial extracellular solute-binding protein, putative, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG.
TO BE PUBLISHED
|
|
4HTF
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase from Escherichia coli in complex with S-adenosylmethionine.
To be Published
|
|
4HVN
 
 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 in complex with Trimethylamine. | | Descriptor: | N,N-dimethylmethanamine, hypothetical protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
4HUS
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with virginiamycin M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
4HN3
 
 | | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e
To be Published
|
|
4I62
 
 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
4HUR
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus in complex with acetyl coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Evdokimova, E, Wawrzak, Z, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
