6GV8
 
 | |
8OEL
 
 | | Condensed RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (8.24 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
6O9R
 
 | |
5HD3
 
 | |
6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8P34
 
 | | Tau filaments extracted from human brain with the DeltaK281 mutation in MAPT | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Garringer, H.J, Klingstedt, T, Masuda-Suzukake, M, Murrell, J.R, Vidal, R, Scheres, S.H.W, Goedert, M, Ghetti, B, Newell, K.L. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Mutation ∆K281 in MAPT causes Pick's disease.
Acta Neuropathol, 146, 2023
|
|
6QQM
 
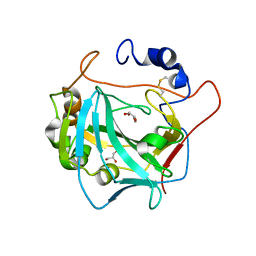 | | Crystal structure of the alpha carbonic anhydrase from Schistosoma mansoni | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Putative carbonic anhydrase, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
7A49
 
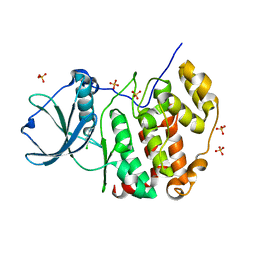 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
8PKF
 
 | |
3I35
 
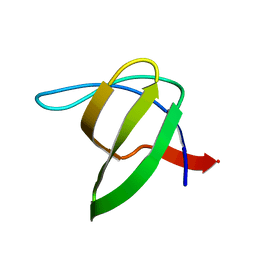 | | Human SH3 domain of protein LASP1 | | Descriptor: | LIM and SH3 domain protein 1 | | Authors: | Siponen, M.I, Roos, A.K, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Kragh Nielsen, T, Moche, M, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human SH3 domain of protein LASP1
TO BE PUBLISHED
|
|
6QYH
 
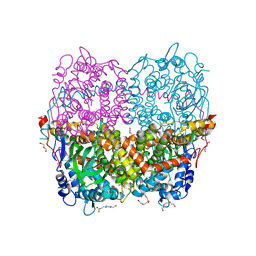 | | Structure of Apo HPAB from E.coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, CALCIUM ION, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
8RHJ
 
 | | Yeast 20S proteasome in complex with a macrocyclic oxindole epoxyketone (compound 5) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Macrocyclic oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
5NXN
 
 | |
6Z9M
 
 | | Pseudoatomic model of the pre-fusion conformation of glycoprotein B of Herpes simplex virus 1 | | Descriptor: | Envelope glycoprotein B | | Authors: | Vollmer, B, Prazak, V, Vasishtan, D, Jefferys, E.E, Hernandez-Duran, A, Vallbracht, M, Klupp, B, Mettenleiter, T.C, Backovic, M, Rey, F.A, Topf, M, Gruenewald, K. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | The prefusion structure of herpes simplex virus glycoprotein B.
Sci Adv, 6, 2020
|
|
3IHJ
 
 | | Human alanine aminotransferase 2 in complex with PLP | | Descriptor: | Alanine aminotransferase 2, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutamate pyruvate transaminase 2
To be Published
|
|
6ZJK
 
 | |
5LH3
 
 | | High dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
7ZQK
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ3
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
5LN8
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica, in complex with galactose | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA, beta-D-galactopyranose | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LND
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LI4
 
 | | bacteriophage phi812K1-420 tail sheath protein after contraction | | Descriptor: | tail sheath protein | | Authors: | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | Deposit date: | 2016-07-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
