4PHX
 
 | |
4PH8
 
 | |
8VPG
 
 | |
8VPI
 
 | |
8VPH
 
 | |
4OR1
 
 | | Structure and mechanism of fibronectin binding and biofilm formation of enteroaggregative Escherischia coli AAF fimbriae | | Descriptor: | ACETATE ION, Invasin homolog AafB, Major fimbrial subunit of aggregative adherence fimbria II AafA chimeric construct, ... | | Authors: | Lee, W.-C, Garnett, J.A, Yang, Y, Matthews, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
1DU0
 
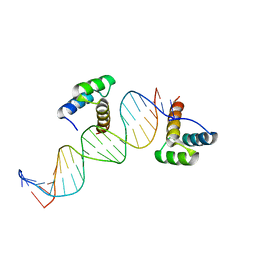 | | ENGRAILED HOMEODOMAIN Q50A VARIANT DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*GP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*CP*CP*TP*AP*A)-3'), ENGRAILED HOMEODOMAIN | | Authors: | Grant, R.A, Rould, M.A, Klemm, J.D, Pabo, C.O. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 --> ala) complex at 2.0 A.
Biochemistry, 39, 2000
|
|
1MNM
 
 | |
5Z55
 
 | |
6JLR
 
 | |
6ILZ
 
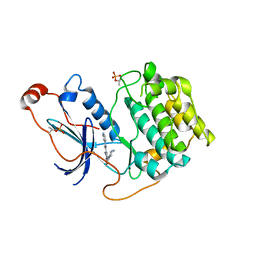 | | Crystal structure of PKCiota in complex with inhibitor | | Descriptor: | 2-amino-5-[3-(piperazin-1-yl)phenyl]-N-(pyridin-4-yl)pyridine-3-carboxamide, Protein kinase C iota type | | Authors: | Baburajendran, N, Hill, J. | | Deposit date: | 2018-10-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.261 Å) | | Cite: | Fragment-based Discovery of a Small-Molecule Protein Kinase C-iota Inhibitor Binding Post-kinase Domain Residues.
Acs Med.Chem.Lett., 10, 2019
|
|
8HZ5
 
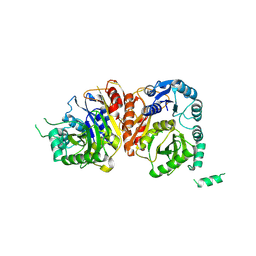 | |
8HZ4
 
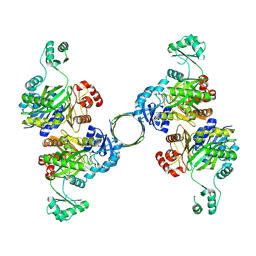 | |
6KAG
 
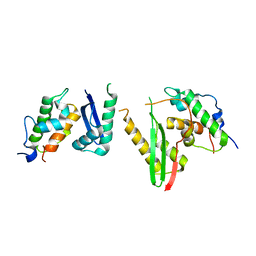 | | Crystal structure of the SMARCB1/SMARCC2 subcomplex | | Descriptor: | SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Chen, G, Zhou, H, Giancotti, F.G, Long, J. | | Deposit date: | 2019-06-22 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | A heterotrimeric SMARCB1-SMARCC2 subcomplex is required for the assembly and tumor suppression function of the BAF chromatin-remodeling complex.
Cell Discov, 6, 2020
|
|
8IQY
 
 | |
8IQV
 
 | |
8IQZ
 
 | |
8IQW
 
 | |
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7CC2
 
 | |
7DT2
 
 | |
7DXL
 
 | |
7VFJ
 
 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7EDR
 
 | |
