5AIW
 
 | |
7NN6
 
 | |
7NMB
 
 | |
1JI9
 
 | |
8OZZ
 
 | |
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
2H3C
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADL
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2JMY
 
 | | Solution structure of CM15 in DPC micelles | | Descriptor: | CM15 | | Authors: | Respondek, M, Madl, T, Goebl, C, Golser, R, Zangger, K. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves
J.Am.Chem.Soc., 129, 2007
|
|
2M64
 
 | | 1H, 13C and 15N Chemical Shift Assignments for Phl p 5a | | Descriptor: | Phlp5 | | Authors: | Goebl, C, Focke, M, Schrank, E, Madl, T, Kosol, S, Madritsch, C, Flicker, S, Valenta, R, Zangger, K, Tjandra, N. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexible IgE epitope-containing domains of Phl p 5 cause high allergenic activity.
J. Allergy Clin. Immunol., 140, 2017
|
|
2KLF
 
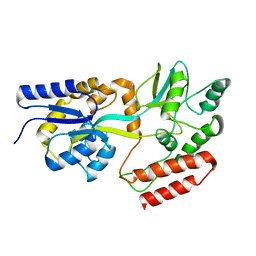 | | PERE NMR structure of maltodextrin-binding protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2KLG
 
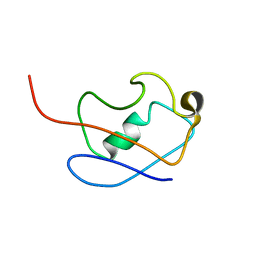 | | PERE NMR structure of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6AEM
 
 | | Crystal structure of the PKD1 domain of Vibrio anguillarum Epp | | Descriptor: | CALCIUM ION, PKD domain, ZINC ION | | Authors: | Ma, Q, Li, P. | | Deposit date: | 2018-08-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | Structural basis for specific calcium binding by the polycystic-kidney-disease domain of Vibrio anguillarum protease Epp
Biochem. Biophys. Res. Commun., 505, 2018
|
|
7DLV
 
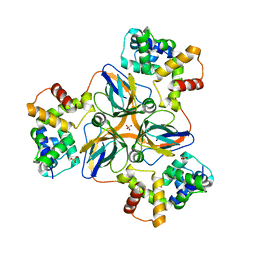 | | shrimp dUTPase in complex with Stl | | Descriptor: | CALCIUM ION, Orf20, SULFATE ION, ... | | Authors: | Ma, Q, Wang, F. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of staphylococcal Stl inhibition on a eukaryotic dUTPase.
Int.J.Biol.Macromol., 184, 2021
|
|
7C4A
 
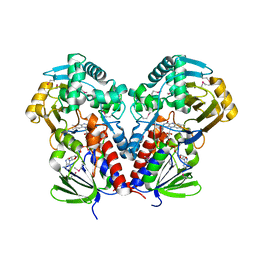 | | nicA2 with cofactor FAD | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
6FY4
 
 | |
6GHW
 
 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
3G7Z
 
 | | CcdB dimer in complex with two C-terminal CcdA domains | | Descriptor: | Cytotoxic protein ccdB, Protein ccdA | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-02-11 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
3HPW
 
 | | CcdB dimer in complex with one C-terminal CcdA domain | | Descriptor: | Cytotoxic protein ccdB, DI(HYDROXYETHYL)ETHER, Protein ccdA, ... | | Authors: | De Jonge, N, Loris, R, Garcia-Pino, A, Buts, L. | | Deposit date: | 2009-06-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Rejuvenation of CcdB-Poisoned Gyrase by an Intrinsically Disordered Protein Domain.
Mol.Cell, 35, 2009
|
|
3JRZ
 
 | | CcdBVfi-FormII-pH5.6 | | Descriptor: | CcdB | | Authors: | De Jonge, N, Buts, L, Loris, R. | | Deposit date: | 2009-09-09 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
3JSC
 
 | | CcdBVfi-FormI-pH7.0 | | Descriptor: | CcdB, SULFATE ION | | Authors: | De Jonge, N, Buts, L, Loris, R. | | Deposit date: | 2009-09-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
2MRN
 
 | |
2MRU
 
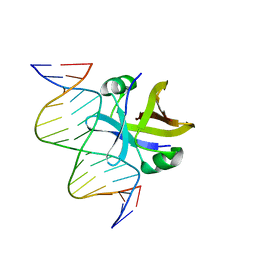 | | Structure of truncated EcMazE-DNA complex | | Descriptor: | Antitoxin MazE, DNA (5'-D(*CP*GP*TP*GP*AP*TP*AP*TP*AP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*CP*TP*AP*TP*AP*TP*AP*TP*CP*AP*CP*G)-3') | | Authors: | Zorzini, V, Buts, L, Loris, R, van Nuland, N. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding.
Nucleic Acids Res., 43, 2015
|
|
