6BIT
 
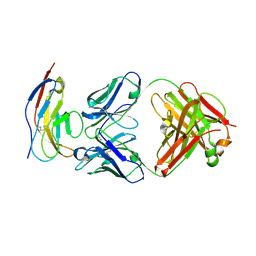 | | SIRPalpha antibody complex | | Descriptor: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | Deposit date: | 2017-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7O1H
 
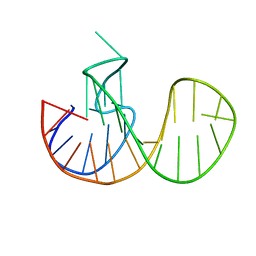 | |
6ZL2
 
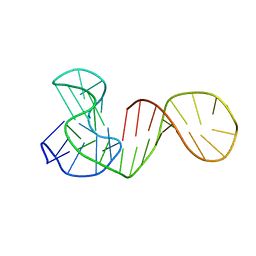 | |
7PNE
 
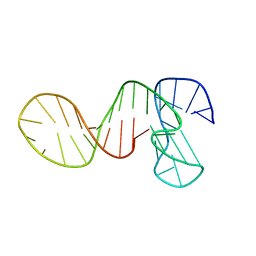 | |
7PNG
 
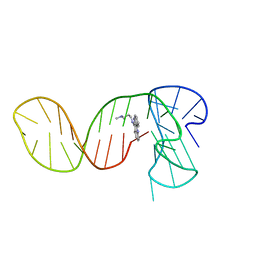 | |
8QKX
 
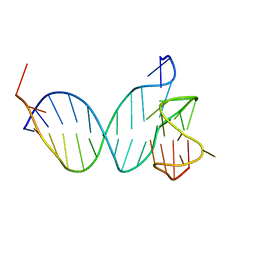 | |
5MCR
 
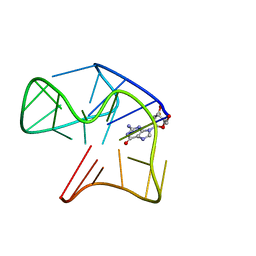 | | Quadruplex with flipped tetrad formed by an artificial sequence | | Descriptor: | Artificial quadruplex with propeller, diagonal and lateral loop | | Authors: | Dickerhoff, J, Haase, L, Langel, W, Weisz, K. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes.
ACS Chem. Biol., 12, 2017
|
|
8R6H
 
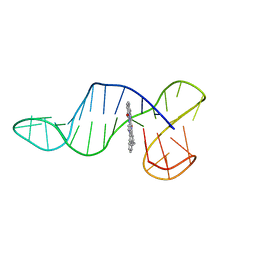 | |
8RW2
 
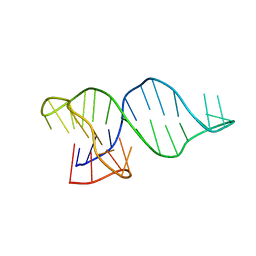 | |
8S1W
 
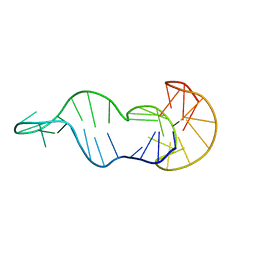 | |
8RZX
 
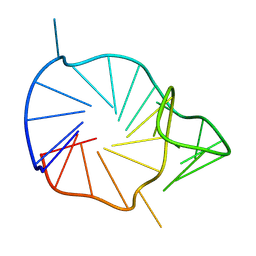 | |
8R6D
 
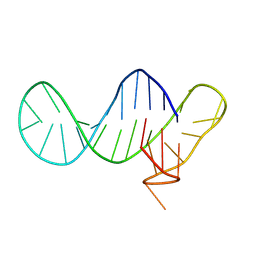 | |
8R6G
 
 | |
5MBR
 
 | |
8R4E
 
 | |
8R4W
 
 | |
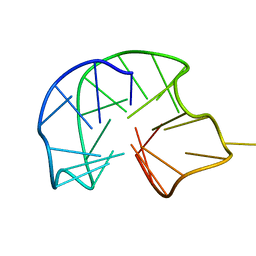
7ATZ
 
 | |
7ZEM
 
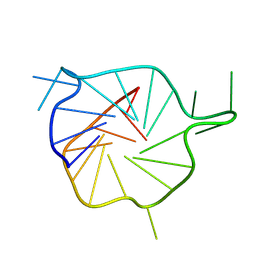 | |
7ZEO
 
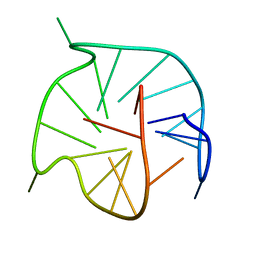 | |
7ZEK
 
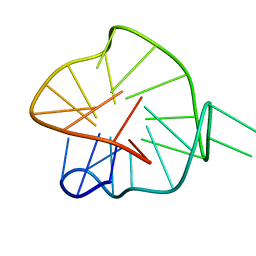 | |
6ZTE
 
 | |
8ABD
 
 | |
8ABN
 
 | |
6F4Z
 
 | |
6FFR
 
 | | DNA-RNA Hybrid Quadruplex with Flipped Tetrad | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*GP*GP*AP*TP*GP*GP*GP*AP*CP*AP*CP*AP*GP*GP*GP*GP*AP*C)-R(P*G)-D(P*GP*G)-3') | | Authors: | Haase, L, Dickerhoff, J, Weisz, K. | | Deposit date: | 2018-01-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DNA-RNA Hybrid Quadruplexes Reveal Interactions that Favor RNA Parallel Topologies.
Chemistry, 24, 2018
|
|
