8BDG
 
 | | Tubulin-taxane-2b complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDF
 
 | | Tubulin-taxane-2a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
8BDE
 
 | | Tubulin-baccatin III complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Lucena-Agell, D, Ma, Y, Estevez-Gallego, J, Li, S, Bargsten, K, Altmann, K.H, Gaillard, N, Kamimura, S, Muehlethaler, T, Gago, F, Oliva, M.A, Steinmetz, M.O, Fang, W.S, Diaz, J.F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural insight into the stabilization of microtubules by taxanes.
Elife, 12, 2023
|
|
6E5I
 
 | |
1F47
 
 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1F46
 
 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1F42
 
 | | THE P40 DOMAIN OF HUMAN INTERLEUKIN-12 | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, INTERLEUKIN-12 BETA CHAIN, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yoon, C, Johnston, S.C, Tang, J, Tobin, J.F, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12.
EMBO J., 19, 2000
|
|
1F7L
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH COENZYME A AT 1.5A | | Descriptor: | CALCIUM ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F7T
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE AT 1.8A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F80
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH HOLO-(ACYL CARRIER PROTEIN) | | Descriptor: | ACYL CARRIER PROTEIN, HOLO-(ACYL CARRIER PROTEIN) SYNTHASE, SODIUM ION | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
6E5J
 
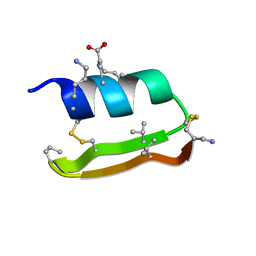 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, beta3 helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, beta3 helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
6E5H
 
 | |
1MUS
 
 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
2Y8K
 
 | | Structure of CtGH5-CBM6, an arabinoxylan-specific xylanase. | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL, ... | | Authors: | Firbank, S.J, Correia, M.A, Mazumder, K, Bras, J.L, Zhu, Y, Lewis, R.J, York, W.S, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and Function of an Arabinoxylan-Specific Xylanase.
J.Biol.Chem., 286, 2011
|
|
1MX6
 
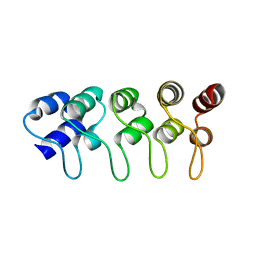 | | Structure of p18INK4c (F92N) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MUH
 
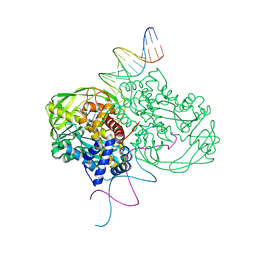 | | CRYSTAL STRUCTURE OF TN5 TRANSPOSASE COMPLEXED WITH TRANSPOSON END DNA | | Descriptor: | DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Davies, D.R, Goryshin, I.Y, Reznikoff, W.S, Rayment, I. | | Deposit date: | 2002-09-23 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the Tn5 synaptic complex transposition intermediate.
Science, 289, 2000
|
|
1MX4
 
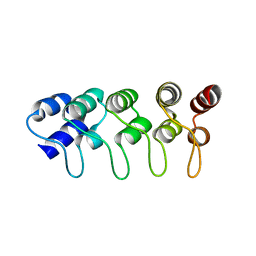 | | Structure of p18INK4c (F82Q) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
2YJ1
 
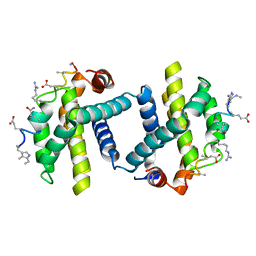 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
4TL1
 
 | | GCN4-p1 with mutation to 1-Aminocyclohexanecarboxylic acid at residue 10 | | Descriptor: | General control protein GCN4, ISOPROPYL ALCOHOL | | Authors: | Tavenor, N.A, Silva, K.I, Saxena, S, Horne, W.S. | | Deposit date: | 2014-05-28 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origins of Structural Flexibility in Protein-Based Supramolecular Polymers Revealed by DEER Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
6UCO
 
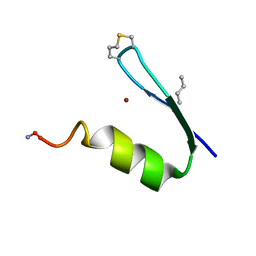 | |
6UCP
 
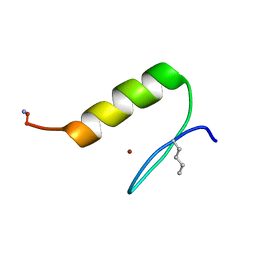 | |
6WYA
 
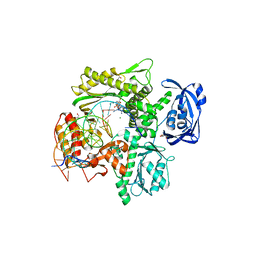 | | RTX (Reverse Transcription Xenopolymerase) in complex with a DNA duplex and dAMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WYB
 
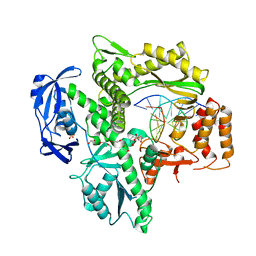 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3BBZ
 
 | | Structure of the nucleocapsid-binding domain from the mumps virus phosphoprotein | | Descriptor: | BROMIDE ION, FORMIC ACID, P protein | | Authors: | Kingston, R.L, Gay, L.S, Baase, W.S, Matthews, B.W. | | Deposit date: | 2007-11-11 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization
J.Mol.Biol., 379, 2008
|
|
3S84
 
 | | Dimeric apoA-IV | | Descriptor: | Apolipoprotein A-IV, SULFATE ION | | Authors: | Deng, X, Davidson, W.S, Thompson, T.B. | | Deposit date: | 2011-05-27 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Dimeric Apolipoprotein A-IV and Its Mechanism of Self-Association.
Structure, 20, 2012
|
|
