7B87
 
 | | Notum-Fragment074 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B86
 
 | | Notum-Fragment067 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8D
 
 | | Notum-Fragment 151 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B4X
 
 | | Notum complex with ARUK3002697 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B89
 
 | | Notum-Fragment077 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
6ZVL
 
 | | ARUK3000263 complex with Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-chloranyl-3-(trifluoromethyl)phenyl]-3~{H}-1,3,4-oxadiazol-2-one, ... | | Authors: | Zhao, Y, Ruza, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
6ZUV
 
 | | Notum fragment 286 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
8PXC
 
 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
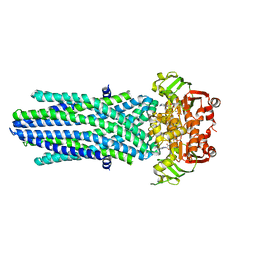 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PWN
 
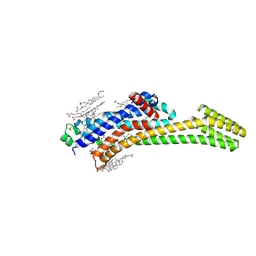 | | Structure of A2A adenosine receptor A2AR-StaR2-bRIL, solved at wavelength 2.75 A | | Descriptor: | Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, OLEIC ACID, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Moraes, I, Wagner, A. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXK
 
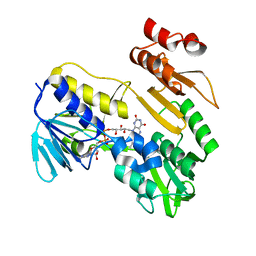 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX1
 
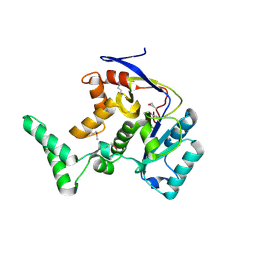 | | Structure of salmonella effector SseK3, solved at wavelength 2.75 A | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C.M, Esposito, D, Rittinger, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX7
 
 | | Structure of Bacterial Multidrug Efflux transporter AcrB, solved at wavelength 3.02 A | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PYZ
 
 | | Structure of Ompk36GD from Klebsiella pneumonia, solved at wavelength 4.13 A | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OmpK36 | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Kwong, H, Beis, K, Wagner, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX5
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX4
 
 | | Structure of the PAS domain code by the LIC_11128 gene from Leptospira interrogans serovar Copenhageni Fiocruz, solved at wavelength 3.09 A | | Descriptor: | Diguanylate cyclase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Guzzo, C.R, Owens, R.J, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXJ
 
 | | Structure of Whitewater Arroyo virus GP1 glycoprotein, solved at wavelength 2.75 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Glycoprotein G1, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bowden, T.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXL
 
 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 1.37 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX0
 
 | | Structure of ribonuclease A, solved at wavelength 2.75 A | | Descriptor: | L-URIDINE-5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXH
 
 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PZ5
 
 | | Structure of ThcOx, solved at wavelength 3.099 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, SagB-type dehydrogenase domain protein | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A. | | Deposit date: | 2023-07-27 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7B99
 
 | | Notum Fragment 283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8U
 
 | | Notum-Fragment 201 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(piperidin-1-yl)-1,2,5-oxadiazol-3-amine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCC
 
 | | Notum Fragment 705 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-pyrrol-1-ylphenyl)morpholine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BD2
 
 | | Notum Fragment 810 | | Descriptor: | DIMETHYL SULFOXIDE, N-[2-(4-hydroxyphenyl)ethyl]pyridine-2-carboxamide, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
