8J0X
 
 | | Crystal structure of horse spleen L-ferritin at -20deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
8J10
 
 | | Crystal structure of horse spleen L-ferritin at -180deg Celsius cooled from -20deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
8J0U
 
 | | Crystal structure of horse spleen L-ferritin A115T mutant at -180deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
8J16
 
 | | Crystal structure of horse spleen L-ferritin at -80deg Celsius. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Maity, B, Tian, J, Abe, S, Ueno, T. | | Deposit date: | 2023-04-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic-Level Insights into a Unique Semi-Clathrate Hydrate Formed in a Confined Environment of Porous Protein Crystal.
Cryst.Growth Des., 23, 2023
|
|
1FP4
 
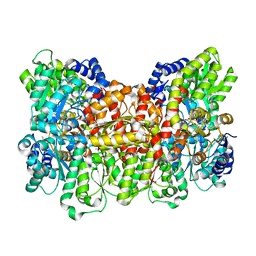 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | Deposit date: | 2000-08-30 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
6UFX
 
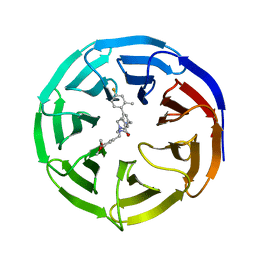 | | WD repeat-containing protein 5 complexed with N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide (compound 13) | | Descriptor: | N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Rietz, T.A, Fesik, S.W, Zhao, B. | | Deposit date: | 2019-09-25 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
J.Med.Chem., 63, 2020
|
|
6UCS
 
 | |
8RKG
 
 | | Crystal structure of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICINE, ... | | Authors: | Nishio, S, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8E9F
 
 | |
8XG2
 
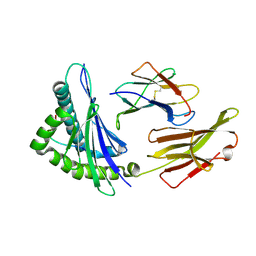 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
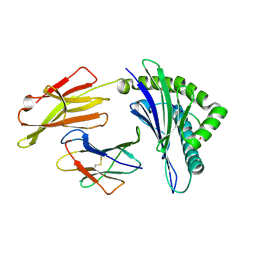 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
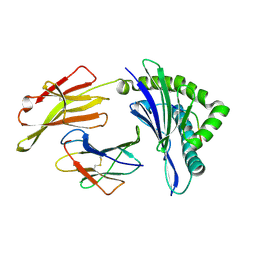 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
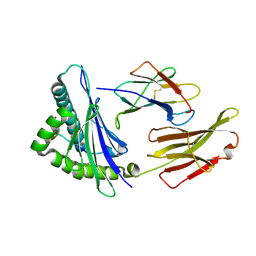 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
7OYH
 
 | |
7OYF
 
 | |
7OY3
 
 | | Crystal structure of depupylase Dop in complex with phosphorylated Pup and ADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXV
 
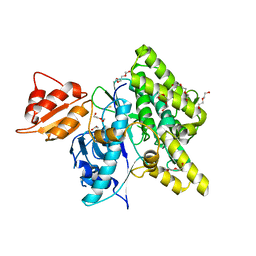 | | Crystal structure of depupylase Dop in the Dop-loop-inserted state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXY
 
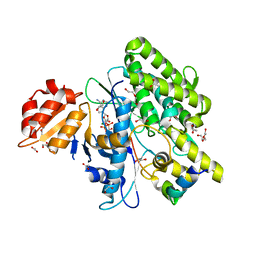 | | Crystal structure of depupylase Dop in complex with Pup and AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7SZE
 
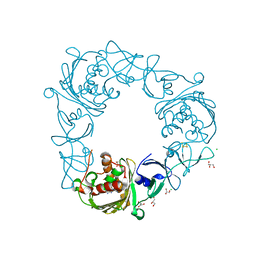 | |
7SZH
 
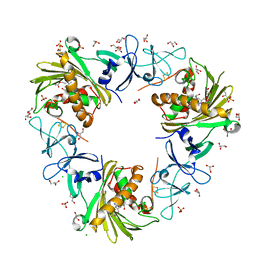 | |
7SZG
 
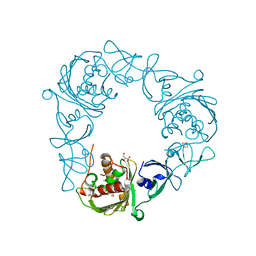 | | Structure of the Rieske Non-heme Iron Oxygenase GxtA Pressurized with Xenon | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Bridwell-Rabb, J, Liu, J. | | Deposit date: | 2021-11-27 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design principles for site-selective hydroxylation by a Rieske oxygenase.
Nat Commun, 13, 2022
|
|
7SZF
 
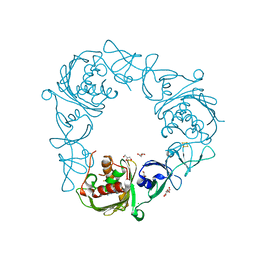 | |
7TXM
 
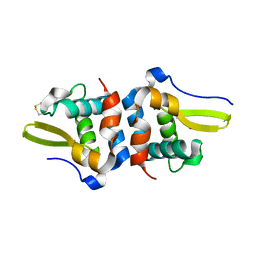 | | Oxidized Structure of RexT | | Descriptor: | HYDROGEN PEROXIDE, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
7TXO
 
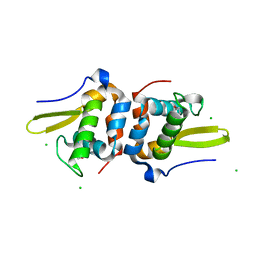 | | Selenomethionine Labeled Structure of RexT | | Descriptor: | CHLORIDE ION, Transcriptional regulator | | Authors: | Bridwell-Rabb, J, Li, B. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT.
Commun Biol, 5, 2022
|
|
