4F8Y
 
 | |
3F7J
 
 | | B.subtilis YvgN | | Descriptor: | NITRATE ION, POTASSIUM ION, YvgN protein | | Authors: | Zhou, Y.F, Lei, J, Liang, Y.H, Su, X.-D. | | Deposit date: | 2008-11-09 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
2FKN
 
 | |
4L22
 
 | |
3CP7
 
 | |
2G0J
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
2G0I
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, hypothetical protein SMU.848 | | Authors: | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
3GAC
 
 | | Structure of mif with HPP | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3GAD
 
 | | Structure of apomif | | Descriptor: | ACETIC ACID, Macrophage migration inhibitory factor-like protein, SULFATE ION | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
3IJT
 
 | | Structural Characterization of SMU.440, a Hypothetical Protein from Streptococcus mutans | | Descriptor: | Putative uncharacterized protein | | Authors: | Nan, J, Brostromer, E, Kristensen, O, Su, X.-D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance
Plos One, 4, 2009
|
|
3OD5
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
2ZBB
 
 | | P43 crystal of DctBp | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID | | Authors: | Zhou, Y.F, Nan, B.Y, Liu, X, Nan, J, Liang, Y.H, Panjikar, S, Ma, Q.J, Wang, Y.P, Su, X.-D. | | Deposit date: | 2007-10-18 | | Release date: | 2008-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | crystal structures of sensory histidine kinase DctBp
to be published
|
|
3BYD
 
 | | Crystal structure of beta-lactamase OXY-1-1 from Klebsiella oxytoca | | Descriptor: | ACETATE ION, Beta-lactamase OXY-1, SULFATE ION | | Authors: | Liang, Y.-H, Wu, S.W, Su, X.-D. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the broadened substrate profile of the extended-spectrum beta-lactamase OXY-1-1 from Klebsiella oxytoca
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3B3D
 
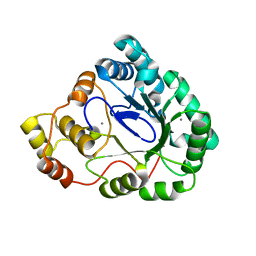 | | B.subtilis YtbE | | Descriptor: | CALCIUM ION, Putative morphine dehydrogenase | | Authors: | Zhou, Y.F, Li, L.F, Liang, Y.H, Su, X.-D. | | Deposit date: | 2007-10-20 | | Release date: | 2008-10-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3CXP
 
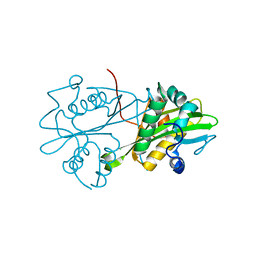 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3CXQ
 
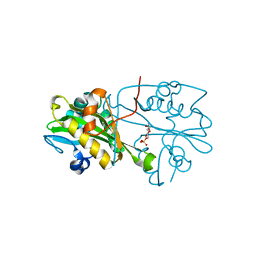 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3LDG
 
 | |
3LDF
 
 | |
3LFJ
 
 | |
3L7P
 
 | |
3L7T
 
 | |
3LCM
 
 | | Crystal structure of Smu.1420 from Streptococcus mutans UA159 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Wang, Z.X, Su, X.-D. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural and biochemical characterization of MdaB from cariogenic Streptococcus mutans reveals an NADPH-specific quinone oxidoreductase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3L8U
 
 | |
3L7N
 
 | |
3LBY
 
 | |
