2D42
 
 | | Crystal structure analysis of a non-toxic crystal protein from Bacillus thuringiensis | | Descriptor: | non-toxic crystal protein | | Authors: | Akiba, T, Higuchi, K, Mizuki, E, Ekino, K, Shin, T, Ohba, M, Kanai, R, Harata, K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to beta-pore-forming toxins
Proteins, 63, 2006
|
|
1N5N
 
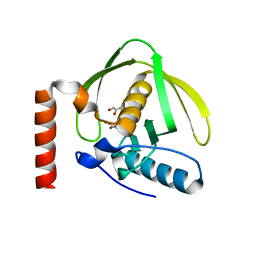 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, Peptide deformylase, ZINC ION | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM4
 
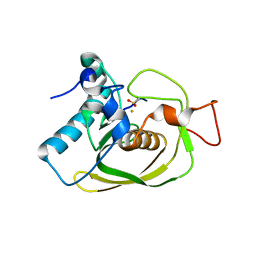 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LM6
 
 | | Crystal Structure of Peptide Deformylase from Streptococcus pneumoniae | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase DEFB | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1QB4
 
 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
6LMI
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
6LMQ
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase catalytic, SULFATE ION, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
6LPG
 
 | | human VASH1-SVBP complex | | Descriptor: | SULFATE ION, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Ikeda, A, Nishino, T. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the tetrameric human vasohibin-1-SVBP complex reveals a variable arm region within the structural core.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8JYH
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid C | | Descriptor: | 7-[5-[(2~{S})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYJ
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid A | | Descriptor: | 7-[5-(2-acetamidoethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYI
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
1N5X
 
 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
1FO4
 
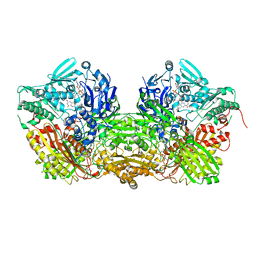 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FIQ
 
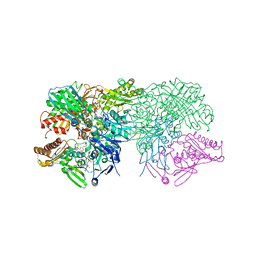 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C7Y
 
 | | E.COLI RUVA-HOLLIDAY JUNCTION COMPLEX | | Descriptor: | DNA (5'-D(P*DAP*DAP*DGP*DTP*DTP*DGP*DGP*DGP*DAP*DTP*DTP*DGP*DT)-3'), DNA (5'-D(P*DCP*DAP*DAP*DTP*DCP*DCP*DCP*DAP*DAP*DCP*DTP*DT)-3'), DNA (5'-D(P*DCP*DGP*DAP*DAP*DTP*DGP*DTP*DGP*DTP*DGP*DTP*DCP*DT)-3'), ... | | Authors: | Ariyoshi, M, Nishino, T, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the holliday junction DNA in complex with a single RuvA tetramer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2E3T
 
 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
3BDJ
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | Descriptor: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
1O5P
 
 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
1QS4
 
 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5JHT
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region crystallized with potassium sodium tartrate | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Influence of Precipitants on Molecular Arrangements and Space Groups of Protein Crystals
Cryst.Growth Des., 17, 2017
|
|
2RQH
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
2RQG
 
 | | Structure of GSPT1/ERF3A-PABC | | Descriptor: | G1 to S phase transition 1, Polyadenylate-binding protein 1 | | Authors: | Osawa, M, Nakanishi, T, Hosoda, N, Uchida, S, Hoshino, T, Katada, I, Shimada, I. | | Deposit date: | 2009-05-08 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic Translation Termination Factor Gspt/Erf3 Recognizes Pabp with Chemical Exchange Using Two Overlapping Motifs
To be Published
|
|
5JHV
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region crystallized with polyethylene glycol 8000 | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Influence of Precipitants on Molecular Arrangements and Space Groups of Protein Crystals
Cryst.Growth Des., 17, 2017
|
|
