8EUA
 
 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3X2W
 
 | | Michaelis complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X2V
 
 | | Michaelis-like complex of cAMP-dependent Protein Kinase Catalytic Subunit | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, MAGNESIUM ION, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
3X2U
 
 | | Michaelis-like initial complex of cAMP-dependent Protein Kinase Catalytic Subunit. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Substrate Peptide, ... | | Authors: | Das, A, Langan, P, Gerlits, O, Kovalevsky, A.Y, Heller, W.T. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Kinase A Catalytic Subunit Primed for Action: Time-Lapse Crystallography of Michaelis Complex Formation.
Structure, 23, 2015
|
|
4QDW
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE5
 
 | |
4QE4
 
 | |
4QEE
 
 | |
4QEH
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE1
 
 | |
5TOQ
 
 | | High resolution crystal structure of AAT | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5TOR
 
 | | Crystal structure of AAT D222T mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5TOT
 
 | |
5VJZ
 
 | | Joint X-ray/neutron structure of aspartate aminotransferase with alpha-methyl-aspartate at pH 7.5 | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, Aspartate aminotransferase, cytoplasmic | | Authors: | Dajnowicz, S, Kovalevsky, A.Y, Mueser, T.C. | | Deposit date: | 2017-04-20 | | Release date: | 2017-11-01 | | Last modified: | 2022-03-16 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct visualization of critical hydrogen atoms in a pyridoxal 5'-phosphate enzyme.
Nat Commun, 8, 2017
|
|
5VK7
 
 | |
5VNS
 
 | | M.tb Antigen 85C Acyl-Enzyme Intermediate with Tetrahydrolipstatin | | Descriptor: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ... | | Authors: | Goins, C.M, Ronning, D.R. | | Deposit date: | 2017-05-01 | | Release date: | 2018-01-24 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mycolyltransferase fromMycobacterium tuberculosisin covalent complex with tetrahydrolipstatin provides insights into antigen 85 catalysis.
J. Biol. Chem., 293, 2018
|
|
5TON
 
 | | Crystal structure of AAT H143L mutant | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Mueser, T.C, Dajnowicz, S, Kovalevsky, A. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase.
J. Biol. Chem., 292, 2017
|
|
5ZO0
 
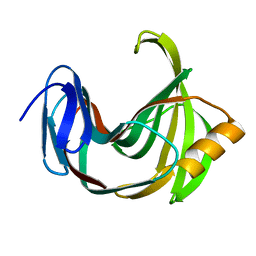 | | Neutron structure of xylanase at pD5.4 | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Wan, Q, Li, Z.H. | | Deposit date: | 2018-04-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.648 Å) | | Cite: | Neutron structure of xylanase at pD5.4
To be published
|
|
5ZKZ
 
 | |
5ZIW
 
 | |
5ZII
 
 | |
2UZQ
 
 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
