5TVZ
 
 | | Solution NMR structure of Saccharomyces cerevisiae Pom152 Ig-like repeat, residues 718-820 | | Descriptor: | Nucleoporin POM152 | | Authors: | Dutta, K, Sampathkumar, P, Cowburn, D, Almo, S.C, Rout, M.P, Fernandez-Martinez, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure, 25, 2017
|
|
5LPV
 
 | |
6ZK7
 
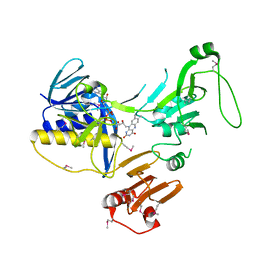 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
5C9F
 
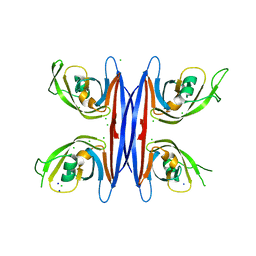 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5C9B
 
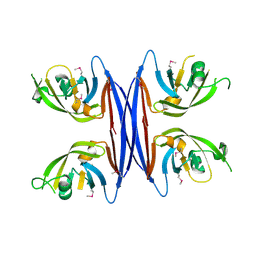 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5C9D
 
 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3OG9
 
 | | Structure of YahD with Malic acid | | Descriptor: | D-MALATE, protein yahD a copper inducible hydrolase | | Authors: | Martinez Font, J, Mancini, S, Tauberger, E, Moniot, S. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Regulation and structure of YahD, a copper inducible alpha/beta hydrolase of Lactococcus lactis IL1403
Fems Microbiol.Lett., 314, 2011
|
|
4IFQ
 
 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
7ZTZ
 
 | | Crystal structure of mutant AR-LBD (Y764C) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZTX
 
 | | Crystal structure of mutant AR-LBD (F755V) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZTV
 
 | | Crystal structure of mutant AR-LBD (F755L) bound to dihydrotestosterone | | Descriptor: | 1,2-ETHANEDIOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU2
 
 | | Crystal structure of mutant AR-LBD (Q799E) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU1
 
 | | Crystal structure of mutant AR-LBD (V758A) bound to dihydrotestosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
1QKW
 
 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
1QKX
 
 | |
5DPX
 
 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
7P3A
 
 | | N-terminal domain of CGI-99 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
7P3B
 
 | | Human RNA ligase RTCB in complex with GMP and Co(II) | | Descriptor: | ACETATE ION, COBALT (II) ION, FORMIC ACID, ... | | Authors: | Kroupova, A, Ackle, F, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
4JMO
 
 | |
4L5M
 
 | |
4JWL
 
 | |
4JXH
 
 | |
4JMI
 
 | |
