7T4V
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-methylbenzoic acid, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4W
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | (2R)-2-({(2S,3S)-1-[(1H-benzimidazol-2-yl)methyl]-2-phenylpiperidin-3-yl}oxy)-2-(3,5-dichlorophenyl)ethan-1-ol, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4T
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4U
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 3-amino-4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)benzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
4CLC
 
 | | Crystal structure of Ybr137w protein | | Descriptor: | UPF0303 PROTEIN YBR137W | | Authors: | Yeh, Y.-H, Lin, T.-W, Lin, C.-Y, Hsiao, C.-D. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterization of Ybr137Wp Implicate its Involvement in the Targeting of Tail-Anchored Proteins to Membranes.
Mol.Cell.Biol., 34, 2014
|
|
2M97
 
 | |
6M84
 
 | | Crystal structure of cKir2.2 force open mutant in complex with PI(4,5)P2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DODECYL-BETA-D-MALTOSIDE, POTASSIUM ION, ... | | Authors: | Lee, S.-J, Ren, F, Yuan, P, Nichols, C.G. | | Deposit date: | 2018-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Atomistic basis of opening and conduction in mammalian inward rectifier potassium (Kir2.2) channels.
J.Gen.Physiol., 152, 2020
|
|
6M85
 
 | |
8TDM
 
 | | Cryo-EM structure of AtMSL10-K539E | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
8TDJ
 
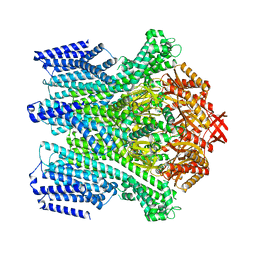 | |
8TDL
 
 | |
8TDK
 
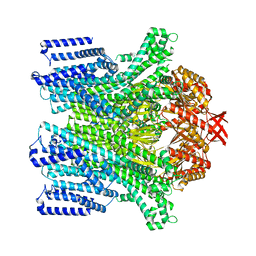 | | Cryo-EM structure of AtMSL10-G556V | | Descriptor: | Mechanosensitive ion channel protein 10 | | Authors: | Zhang, J, Yuan, P. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10.
Nat Commun, 14, 2023
|
|
6UW6
 
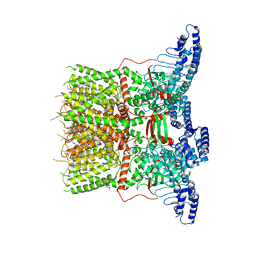 | | Cryo-EM structure of the human TRPV3 K169A mutant determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW9
 
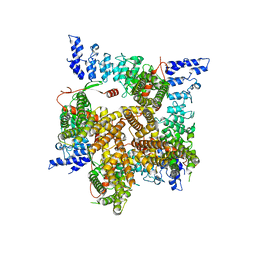 | |
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V6D
 
 | | Cryo-EM structure of human pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW8
 
 | |
6UZY
 
 | | Cryo-EM structure of Xenopus tropicalis pannexin 1 | | Descriptor: | HEXADECANE, Pannexin, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VXN
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 A320V | | Descriptor: | DODECANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6BBJ
 
 | | Xenopus Tropicalis TRPV4 | | Descriptor: | Transient receptor potential cation channel, subfamily V, member 4 | | Authors: | Deng, Z, Hite, R.K, Yuan, P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6VXP
 
 | |
6VXM
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 | | Descriptor: | EICOSANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6GEZ
 
 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6CTD
 
 | |
