9MQT
 
 | | CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Magnesium Condition | | Descriptor: | MAGNESIUM ION, N~4~-(2-aminoethyl)-N~4~-methylpyrimidine-2,4-diamine, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6M19
 
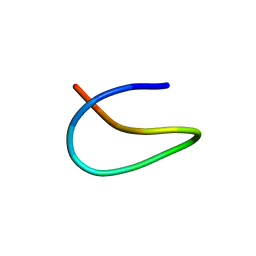 | | Template lasso peptide C24 mutant W14F | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-02-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
3S6T
 
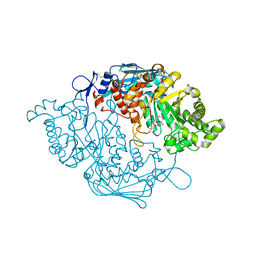 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Yang, Q, Shen, X. | | Deposit date: | 2011-05-26 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases.
Biochem.J., 438, 2011
|
|
3NSN
 
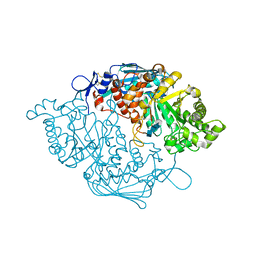 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | Descriptor: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3OZP
 
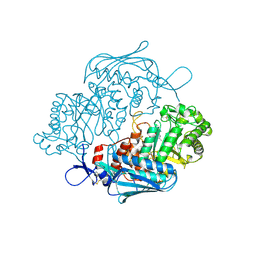 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases
Biochem.J., 438, 2011
|
|
3NSM
 
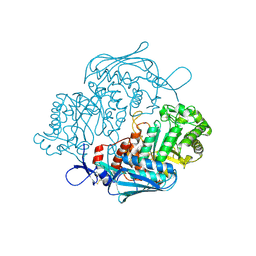 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
4EBY
 
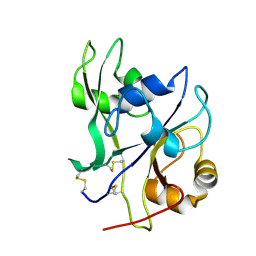 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
4EBZ
 
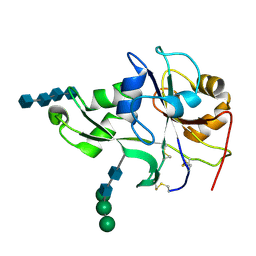 | | Crystal structure of the ectodomain of a receptor like kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZA
 
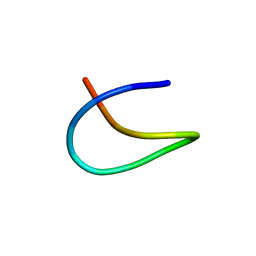 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
8X3Y
 
 | | ThDP-dependent HKA synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BbmA, MAGNESIUM ION, ... | | Authors: | Yu, J.H, Liu, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into two thiamine diphosphate-dependent enzymes and their synthetic applications in carbon-carbon linkage reactions.
Nat.Chem., 2025
|
|
8X3Z
 
 | | ThDP-dependent HKA synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CsmA, MAGNESIUM ION, ... | | Authors: | Yu, J.H, Liu, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into two thiamine diphosphate-dependent enzymes and their synthetic applications in carbon-carbon linkage reactions.
Nat.Chem., 2025
|
|
8X3X
 
 | | ThDP-dependent HKA synthase | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ADENOSINE-5'-DIPHOSPHATE, BbmA, ... | | Authors: | Yu, J.H, Liu, T. | | Deposit date: | 2023-11-14 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into two thiamine diphosphate-dependent enzymes and their synthetic applications in carbon-carbon linkage reactions.
Nat.Chem., 2025
|
|
8XOD
 
 | | ThDP-dependent HKA synthase | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(~{R})-cyclobutyl(oxidanyl)methyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, BbmA-G484F complex with CBOA, ... | | Authors: | Yu, J.H, Liu, T. | | Deposit date: | 2024-01-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into two thiamine diphosphate-dependent enzymes and their synthetic applications in carbon-carbon linkage reactions.
Nat.Chem., 2025
|
|
4D6V
 
 | | Crystal structure of signal transducing protein | | Descriptor: | G PROTEIN BETA SUBUNIT GIB2 | | Authors: | Ero, R, Dimitrova, V.T, Chen, Y, Bu, W, Feng, S, Liu, T, Wang, P, Xue, C, Tan, S.M, Gao, Y.G. | | Deposit date: | 2014-11-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Gib2, a Signal-Transducing Protein Scaffold Associated with Ribosomes in Cryptococcus Neoformans.
Sci.Rep., 5, 2015
|
|
6LK3
 
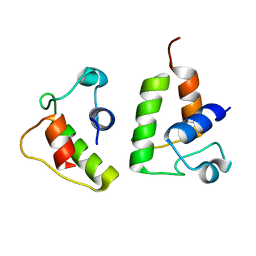 | | The Functional Characterization and Crystal Structure of Type II Peptidyl Carrier Protein ColA1a in Collismycins Biosynthesis | | Descriptor: | Putative free-standing acyl carrier protein | | Authors: | Ma, X.Y, Wang, G.Y, Liu, T, Chi, C.B, Zhang, Z.Y, Yang, D.H, Liu, W, Ma, M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional characterization and crystal structure of type II peptidyl carrier protein ColA1a in collismycins biosynthesis.
Chin.J.Chem., 38, 2020
|
|
3OZO
 
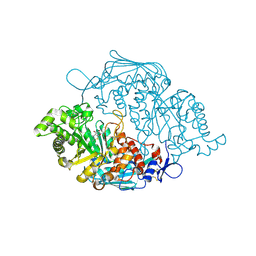 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
5GVX
 
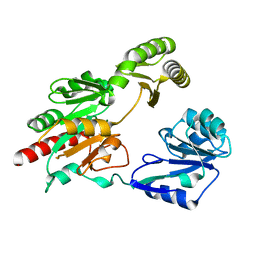 | | Structural insight into dephosphorylation by Trehalose 6-phosphate Phosphatase (OtsB2) from Mycobacterium Tuberculosis | | Descriptor: | MAGNESIUM ION, Trehalose-phosphate phosphatase | | Authors: | Shan, S, Min, H, Liu, T, Jiang, D, Rao, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural insight into dephosphorylation by trehalose 6-phosphate phosphatase (OtsB2) from Mycobacterium tuberculosis.
FASEB J., 30, 2016
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | Authors: | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-20 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
4F7Z
 
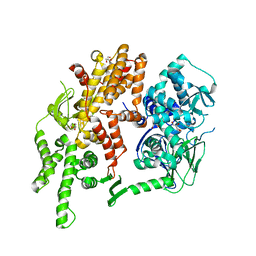 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
