3IBB
 
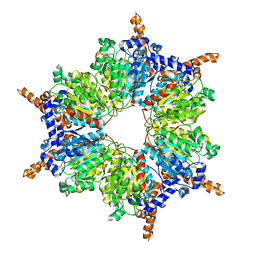 | | Propionyl-CoA Carboxylase Beta Subunit, D422A | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
6X0R
 
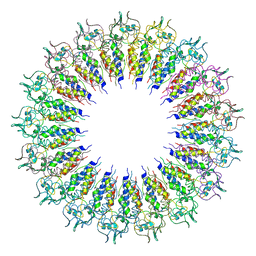 | |
6X0Q
 
 | |
1B35
 
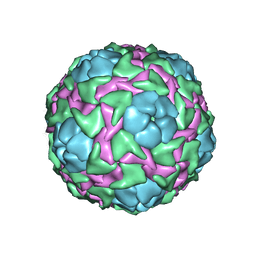 | | CRICKET PARALYSIS VIRUS (CRPV) | | Descriptor: | PROTEIN (CRICKET PARALYSIS VIRUS, VP1), VP2), ... | | Authors: | Tate, J.G, Liljas, L, Scotti, P.D, Christian, P.D, Lin, T.W, Johnson, J.E. | | Deposit date: | 1998-12-17 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cricket paralysis virus: the first view of a new virus family.
Nat.Struct.Biol., 6, 1999
|
|
4REF
 
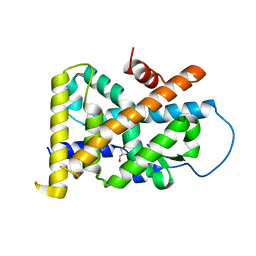 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
8Y7L
 
 | |
4RE8
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
5KWX
 
 | |
5KX2
 
 | |
5KWZ
 
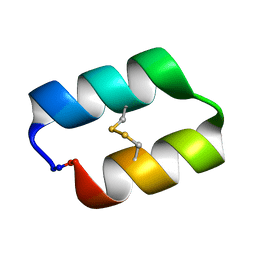 | |
5KX1
 
 | |
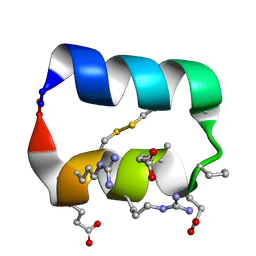
5KX0
 
 | |
5KWP
 
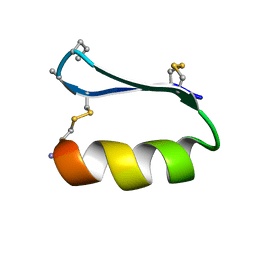 | |
5KVN
 
 | |
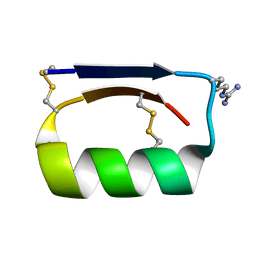
5KWO
 
 | |
5JG9
 
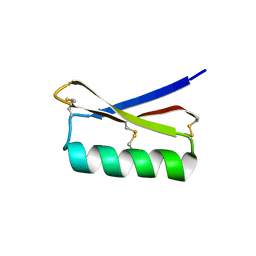 | |
5HUL
 
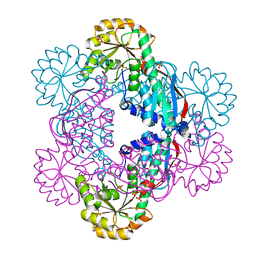 | | Crystal Structure of NadC Deletion Mutant in Cubic Space Group | | Descriptor: | PHOSPHATE ION, Quinolinate phosphoribosyltransferase | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUJ
 
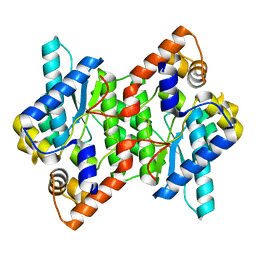 | | Crystal Structure of NadE from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUH
 
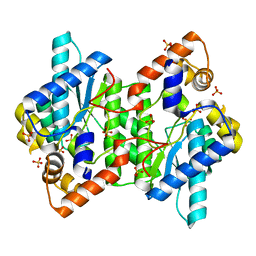 | | Crystal Structure of NadE from Streptococcus pyogenes | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUP
 
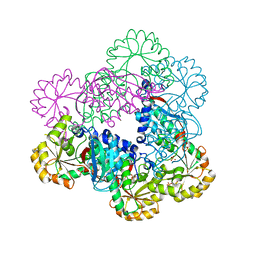 | | Crystal Structure of NadC from Streptococcus pyogenes | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase (Carboxylating), SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUO
 
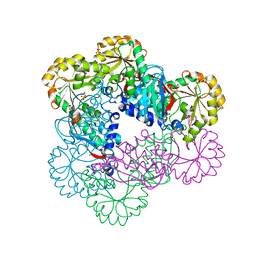 | | Crystal Structure of NadC Deletion Mutant in C2221 Space Group | | Descriptor: | Nicotinate-nucleotide diphosphorylase (Carboxylating), SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
6V1M
 
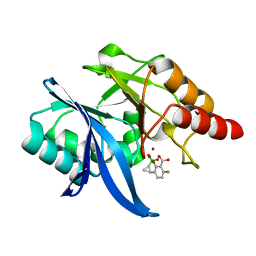 | | Structure of NDM-1 bound to QPX7728 at 1.05 A | | Descriptor: | (1~{a}~{R},7~{b}~{S})-5-fluoranyl-2,2-bis(oxidanyl)-1~{a},7~{b}-dihydro-1~{H}-cyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
6V1J
 
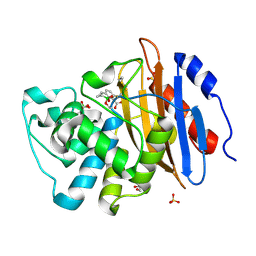 | | Structure of KPC-2 bound to QPX7728 at 1.30 A | | Descriptor: | (1aR,7bS)-5-fluoro-2-hydroxy-1,1a,2,7b-tetrahydrocyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, (1~{a}~{R},7~{b}~{S})-5-fluoranyl-2,2-bis(oxidanyl)-1~{a},7~{b}-dihydro-1~{H}-cyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
6V1P
 
 | | Structure of VIM-2 bound to QPX7728 at 1.20 A | | Descriptor: | (1~{a}~{R},7~{b}~{S})-5-fluoranyl-2,2-bis(oxidanyl)-1~{a},7~{b}-dihydro-1~{H}-cyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
