6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMT
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7O9F
 
 | | Bacillus subtilis Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9I
 
 | | Escherichia coli Ffh in complex with pppGpp | | Descriptor: | Signal recognition particle protein, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9G
 
 | | Escherichia coli Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9H
 
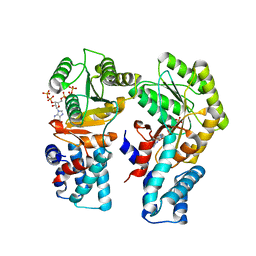 | | Escherichia coli FtsY in complex with pppGpp | | Descriptor: | Signal recognition particle receptor FtsY, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
4C9T
 
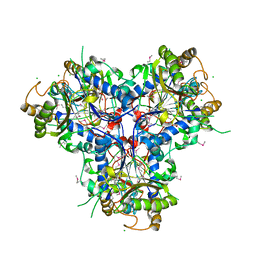 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D06
 
 | | Bacterial chalcone isomerase complexed with naringenin | | Descriptor: | (2E)-3-(4-hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)prop-2-en-1-one, CHALCONE ISOMERASE, CHLORIDE ION, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8B7Z
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Palm, G.J, Hinrichs, W. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7R
 
 | | Bacterial chalcone isomerase with taxifolin chalcone | | Descriptor: | (Z)-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-1-[2,4,6-tris(oxidanyl)phenyl]prop-2-en-1-one, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7U
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
4D4F
 
 | | Mutant P250A of bacterial chalcone isomerase from Eubacterium ramulus | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL | | Authors: | Thomsen, M, Kratzat, H, Hinrichs, W. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus.
Molecules, 27, 2022
|
|
