1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | Authors: | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
1YQ3
 
 | | Avian respiratory complex ii with oxaloacetate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Coenzyme Q10, (2Z,6E,10Z,14E,18E,22E,26Z)-isomer, ... | | Authors: | Huang, L, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
6YL5
 
 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMI
 
 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMJ
 
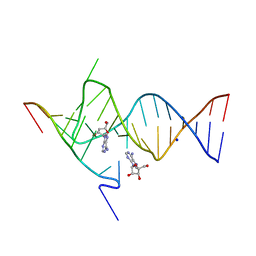 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YLB
 
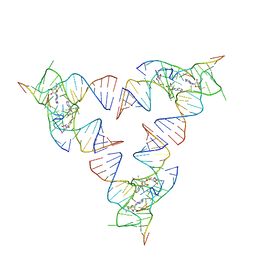 | | Crystal structure of the SAM-SAH riboswitch with SAM | | Descriptor: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YML
 
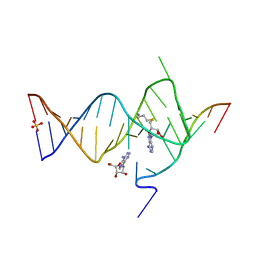 | |
6YMK
 
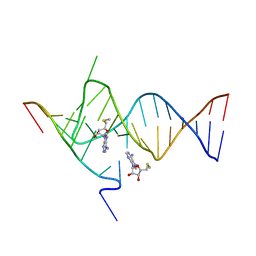 | | Crystal structure of the SAM-SAH riboswitch with AMP | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
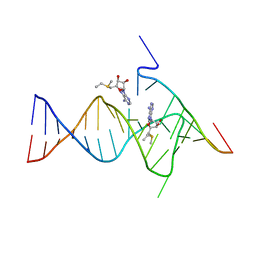
6YMM
 
 | |
5LQT
 
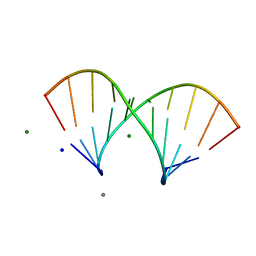 | |
4C4W
 
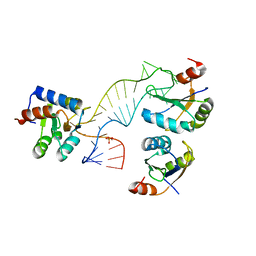 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4C40
 
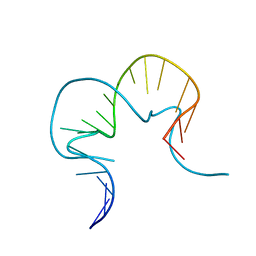 | |
4BW0
 
 | |
1C4Z
 
 | | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY | | Descriptor: | UBIQUITIN CONJUGATING ENZYME E2, UBIQUITIN-PROTEIN LIGASE E3A | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
5FKF
 
 | |
5FK1
 
 | |
5FKH
 
 | |
5G4V
 
 | |
6TF1
 
 | |
6TFF
 
 | |
6TF0
 
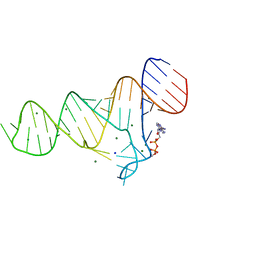 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
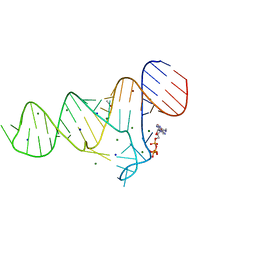 | |
6TB7
 
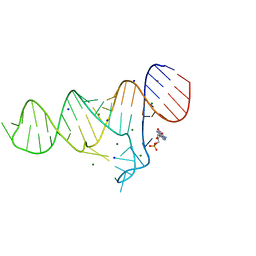 | |
