7ZCG
 
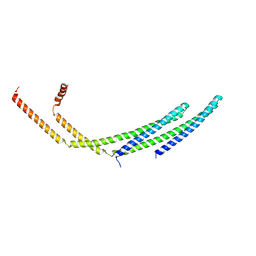 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PQH
 
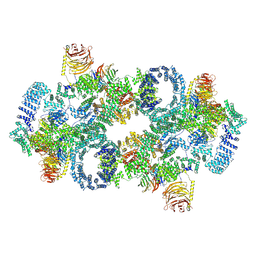 | | Cryo-EM structure of Saccharomyces cerevisiae TOROID (TORC1 Organized in Inhibited Domains). | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 1 subunit KOG1,Target of rapamycin complex 1 subunit Kog1, Target of rapamycin complex subunit LST8 | | Authors: | Felix, J, Prouteau, M, Bourgoint, C, Bonadei, L, Desfosses, A, Gabus, C, Sadian, Y, Savvides, S.N, Gutsche, I, Loewith, R. | | Deposit date: | 2021-09-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | EGOC inhibits TOROID polymerization by structurally activating TORC1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6GGS
 
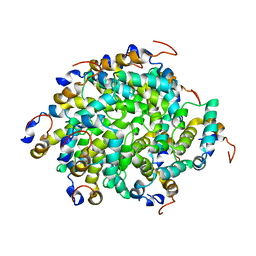 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
4CBW
 
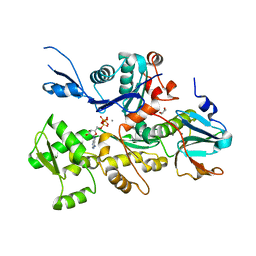 | | Crystal structure of Plasmodium berghei actin I with D-loop from muscle actin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
4CBU
 
 | | Crystal structure of Plasmodium falciparum actin I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
4CBX
 
 | | Crystal structure of Plasmodium berghei actin II | | Descriptor: | ACTIN-2, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
6GK2
 
 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBG
 
 | |
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QBE
 
 | |
8QBB
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QB9
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30) | | Descriptor: | Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8PK7
 
 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-25 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Alphavirus nsP3 organizes into tubular scaffolds essential for infection and the cytoplasmic granule architecture.
Nat Commun, 15, 2024
|
|
8QB8
 
 | |
8QB7
 
 | |
8PHZ
 
 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Alphavirus nsP3 organizes into tubular scaffolds essential for infection and the cytoplasmic granule architecture.
Nat Commun, 15, 2024
|
|
8PHE
 
 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
3QS7
 
 | |
3QS9
 
 | |
5M2N
 
 | |
5JCG
 
 | |
9GNS
 
 | | X-ray structure of Human holo aromatic L-amino acid decarboxylase (AADC) complex with Carbidopa at physiological pH | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CARBIDOPA, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Perduca, M, Bisello, G, Bertoldi, M. | | Deposit date: | 2024-09-04 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
6CVZ
 
 | | Crystal structure of the WD40-repeat of RFWD3 | | Descriptor: | E3 ubiquitin-protein ligase RFWD3, MAGNESIUM ION | | Authors: | DONG, A, LOPPNAU, P, SEITOVA, A, HUTCHINSON, A, TEMPEL, W, WEI, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target highlights in CASP13: Experimental target structures through the eyes of their authors.
Proteins, 87, 2019
|
|
