2FG1
 
 | | Structure of a Protein of Unknown Function from Bacteroides thetaiotaomicron. | | Descriptor: | conserved hypothetical protein BT1257 | | Authors: | Cuff, M.E, Bigelow, L, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a hypothetical protein from Bacteroides thetaiotaomicron.
To be Published
|
|
2FCK
 
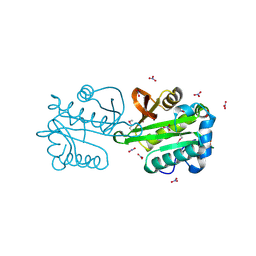 | | Structure of a putative ribosomal-protein-serine acetyltransferase from Vibrio cholerae. | | Descriptor: | GLYCEROL, NITRATE ION, ribosomal-protein-serine acetyltransferase, ... | | Authors: | Cuff, M.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-28 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an acetyltransferase protein from Vibrio cholerae strain N16961.
Proteins, 69, 2007
|
|
4H7N
 
 | |
4HG2
 
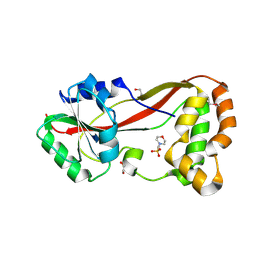 | | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Methyltransferase type 11 | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans.
TO BE PUBLISHED
|
|
4HB7
 
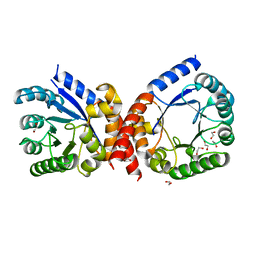 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
4HES
 
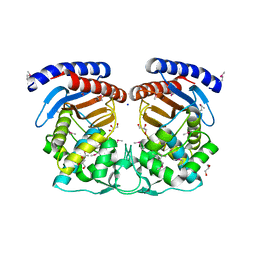 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
4H89
 
 | | The Structure of a GCN5-Related N-Acetyltransferase from Kribbella flavida | | Descriptor: | GCN5-related N-acetyltransferase | | Authors: | Cuff, M.E, Mcknight, S.M, Mack, J.C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of a GCN5-Related N-Acetyltransferase from Kribbella flavida.
TO BE PUBLISHED
|
|
4HBZ
 
 | | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia. | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase, ... | | Authors: | Cuff, M.E, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of Putative Phosphohistidine Phosphatase SixA from Nakamurella multipartitia.
TO BE PUBLISHED
|
|
1XQA
 
 | |
1ZX5
 
 | | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-06 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative mannosephosphate isomerase from Archaeoglobus fulgidus
To be Published
|
|
1ZS7
 
 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
1Y8A
 
 | | Structure of gene product AF1437 from Archaeoglobus fulgidus | | Descriptor: | MAGNESIUM ION, hypothetical protein AF1437 | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of gene product AF1437 from Archaeoglobus fulgidus
To be published
|
|
1Y0B
 
 | |
1Z72
 
 | | Structure of a putative transcriptional regulator from Streptococcus pneumoniae | | Descriptor: | ACETIC ACID, MAGNESIUM ION, transcriptional regulator, ... | | Authors: | Cuff, M.E, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator from Streptococcus pneumoniae
To be Published
|
|
1YB2
 
 | | Structure of a putative methyltransferase from Thermoplasma acidophilum. | | Descriptor: | hypothetical protein Ta0852 | | Authors: | Cuff, M.E, Xu, X, Edwards, A, Savchenko, A, Sanishvili, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-18 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of a putative methyltransferase from Thermoplasma acidophilum.
To be Published
|
|
3H35
 
 | | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2. | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, uncharacterized protein ABO_0056 | | Authors: | Cuff, M.E, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2.
TO BE PUBLISHED
|
|
1PBJ
 
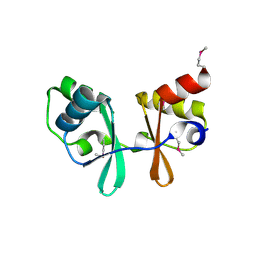 | | CBS domain protein | | Descriptor: | MAGNESIUM ION, hypothetical protein | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-14 | | Release date: | 2003-12-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a hypothetical protein from M. thermautotrophicus reveals a
novel fold and a pseudo 2-fold axis of symmetry
TO BE PUBLISHED
|
|
3H36
 
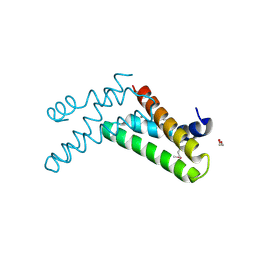 | | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
3HH1
 
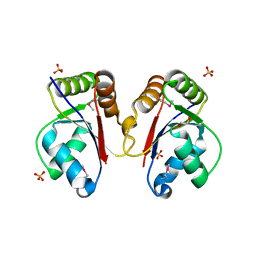 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
3HE0
 
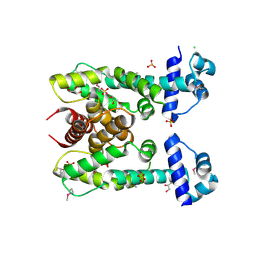 | | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus. | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Hendricks, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Putative Transcriptional Regulator TetR Family Protein from Vibrio parahaemolyticus.
TO BE PUBLISHED
|
|
3IC3
 
 | | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Jedrzejczak, R, Mckinlay, J.B, Harwood, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009
TO BE PUBLISHED
|
|
1PW5
 
 | | putative nagD protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, SULFATE ION, nagD protein, ... | | Authors: | Cuff, M.E, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-30 | | Release date: | 2004-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | putative nagD protein
To be Published
|
|
1OU0
 
 | | precorrin-8X methylmutase related protein | | Descriptor: | precorrin-8X methylmutase related protein | | Authors: | Cuff, M.E, Joachimiak, A, Korolev, S, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-24 | | Release date: | 2003-10-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted precorrin-8x methylmutase from Thermoplasma acidophilum.
Proteins, 58, 2004
|
|
3IJM
 
 | | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale. | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Cuff, M.E, Tesar, C, Samano, S, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a restriction endonuclease-like fold superfamily protein from Spirosoma linguale.
TO BE PUBLISHED
|
|
1Q7H
 
 | | Structure of a Conserved PUA Domain Protein from Thermoplasma acidophilum | | Descriptor: | ZINC ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-18 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a conserved hypothetical protein from T. acidophilum
TO BE PUBLISHED
|
|
