5Z9H
 
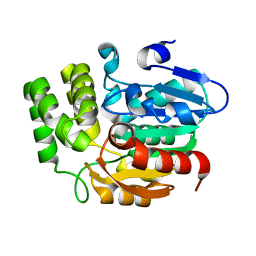 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5FJG
 
 | | The crystal structure of light-driven chloride pump ClR in pH 4.5. | | Descriptor: | ANHYDRORETINOL, CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Light-Driven Chloride Pump Clr in Ph 4.5.
To be Published
|
|
5G2C
 
 | | The crystal structure of light-driven chloride pump ClR (T102D) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2A
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0 with Bromide ion. | | Descriptor: | BROMIDE ION, CHLORIDE PUMPING RHODOPSIN, RETINAL | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G54
 
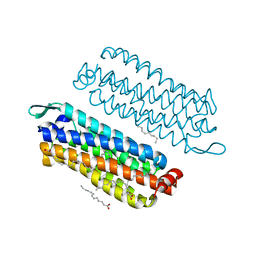 | | The crystal structure of light-driven chloride pump ClR at pH 4.5 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G28
 
 | | The crystal structure of light-driven chloride pump ClR at pH 6.0. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, OLEIC ACID, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
5G2D
 
 | | The crystal structure of light-driven chloride pump ClR (T102N) mutant at pH 4.5. | | Descriptor: | CHLORIDE ION, CHLORIDE PUMP RHODOPSIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Functional Characterization of a Light-Driven Chloride Pump Having an Ntq Motif.
Nat.Commun., 7, 2016
|
|
6KF9
 
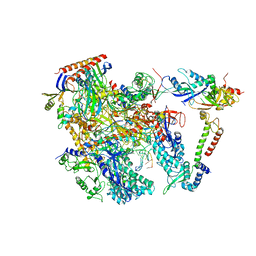 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF3
 
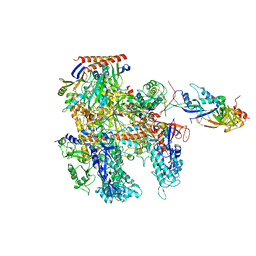 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
1DRV
 
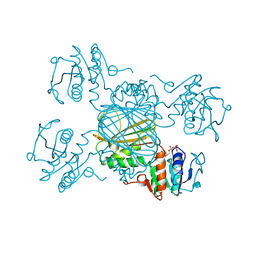 | | ESCHERICHIA COLI DHPR/ACNADH COMPLEX | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1DRU
 
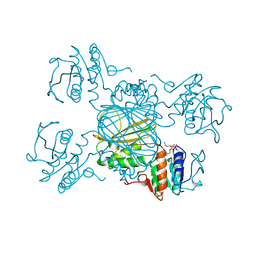 | | ESCHERICHIA COLI DHPR/NADH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1DRW
 
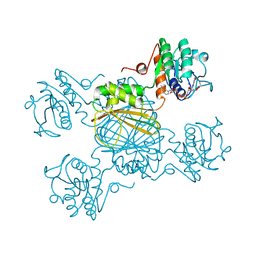 | | ESCHERICHIA COLI DHPR/NHDH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE PURIN-6-OL-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
8E4Y
 
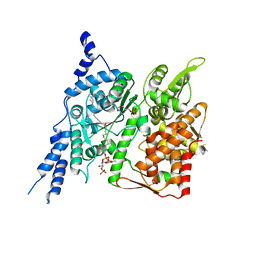 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E50
 
 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, COENZYME A, Glycerol-3-phosphate acyltransferase 1, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6N0S
 
 | | N-terminal domain of Staphylothermus marinus McrB | | Descriptor: | ATPase associated with various cellular activities, AAA_5, SULFATE ION | | Authors: | Hosford, C.J, Niu, Y, Chappie, J.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The N-terminal domain of Staphylothermus marinus McrB shares structural homology with PUA-like RNA binding proteins.
J.Struct.Biol., 211, 2020
|
|
6OWO
 
 | | CRYO-EM STRUCTURE OF PHOSPHORYLATED AP-2 CORE BOUND TO NECAP | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Partlow, E.A, Baker, R.W, Beacham, G.M, Chappie, J.S, Leschziner, A.E, Hollopeter, G. | | Deposit date: | 2019-05-10 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A structural mechanism for phosphorylation-dependent inactivation of the AP2 complex.
Elife, 8, 2019
|
|
6NJX
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with mercury | | Descriptor: | IODIDE ION, MERCURY (II) ION, Xcc_ctr_Hg | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NJW
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with platinum | | Descriptor: | IODIDE ION, MAGNESIUM ION, PLATINUM (II) ION, ... | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NJV
 
 | | C-terminal region of the Xanthomonas campestris pv. campestris OLD protein phased with iodine | | Descriptor: | IODIDE ION, MAGNESIUM ION, Xcc_CTR_I | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
6NK8
 
 | | C-terminal region of the Burkholderia pseudomallei OLD protein | | Descriptor: | Class 2 OLD family nuclease, MAGNESIUM ION | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
1DAP
 
 | | C. GLUTAMICUM DAP DEHYDROGENASE IN COMPLEX WITH NADP+ | | Descriptor: | ACETATE ION, DIAMINOPIMELIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scapin, G, Reddy, S.G, Blanchard, J.S. | | Deposit date: | 1996-07-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of meso-diaminopimelic acid dehydrogenase from Corynebacterium glutamicum.
Biochemistry, 35, 1996
|
|
1DIH
 
 | |
6P0F
 
 | | N-terminal domain of Thermococcus Gammatolerans McrB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, GTPase subunit of restriction endonuclease, ... | | Authors: | Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | The structure of theThermococcus gammatoleransMcrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.
J.Biol.Chem., 295, 2020
|
|
6P0G
 
 | | N-terminal domain of Thermococcus Gammatolerans McrB bound to m5C DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*AP*CP*CP*GP*G)-3'), GTPase subunit of restriction endonuclease | | Authors: | Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of theThermococcus gammatoleransMcrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.
J.Biol.Chem., 295, 2020
|
|
6UT6
 
 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
