3SV2
 
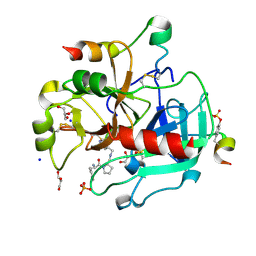 | | Human Thrombin In Complex With UBTHR105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-(pyridin-4-ylmethyl)-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3T8C
 
 | |
3UWJ
 
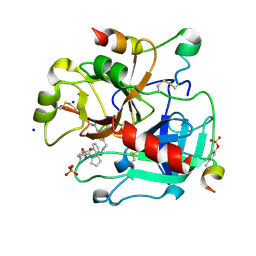 | | Human Thrombin In Complex With MI353 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, N-(benzylsulfonyl)-D-leucyl-N-(4-carbamimidoylbenzyl)-L-prolinamide, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-12-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3U98
 
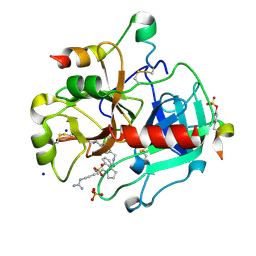 | | Human Thrombin In Complex With MI001 | | Descriptor: | (2S)-1-[(2R)-2-(benzylsulfonylamino)-5-guanidino-pentanoyl]-N-[(4-carbamimidoylphenyl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-10-18 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3T5F
 
 | | Human Thrombin In Complex With MI340 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
3T8G
 
 | | Thermolysin In Complex With UBTLN26 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-08-01 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water makes the difference: rearrangement of water solvation layer triggers non-additivity of functional group contributions in protein-ligand binding.
Chemmedchem, 7, 2012
|
|
3T8D
 
 | |
4H57
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dissecting the hydrophobic effect on the molecular level: the role of water, enthalpy, and entropy in ligand binding to thermolysin.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8AT6
 
 | | Cryo-EM structure of yeast Elp456 subcomplex | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASW
 
 | | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Alanine tRNA, Elongator complex protein 1, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASV
 
 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
3FLF
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, GLYCEROL, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-valyl-L-leucine, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2008-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: binding of phosphonamidate to the S1'-pocket of thermolysin.
Biochim.Biophys.Acta, 1800, 2010
|
|
3FV4
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: binding of phosphonamidate to the S1'-pocket of thermolysin.
Biochim.Biophys.Acta, 1800, 2010
|
|
3FVP
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2009-01-16 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Thermolysin inhibition
To be Published
|
|
5IO7
 
 | |
5IO5
 
 | |
5IO6
 
 | |
4PCZ
 
 | | Crystal structure of a complex between R247G LlFPG mutant and a THF containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Zinc finger oxidation of Fpg/Nei DNA glycosylases by 2-thioxanthine: biochemical and X-ray structural characterization.
Nucleic Acids Res., 42, 2014
|
|
4PDI
 
 | |
4PD2
 
 | | Crystal structure of a complex between a C248GH LlFpg mutant and a THF containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Zinc finger oxidation of Fpg/Nei DNA glycosylases by 2-thioxanthine: biochemical and X-ray structural characterization.
Nucleic Acids Res., 42, 2014
|
|
4PDG
 
 | | Crystal structure of a complex between an inhibited LlFpg and a THF containing DNA | | Descriptor: | 2-sulfanyl-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Castaing, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zinc finger oxidation of Fpg/Nei DNA glycosylases by 2-thioxanthine: biochemical and X-ray structural characterization.
Nucleic Acids Res., 42, 2014
|
|
6IC0
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 4 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-pyrimidin-5-yl-pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6IBX
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 5 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpiperidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
8A0E
 
 | | CryoEM structure of DHS-eIF5A1 complex | | Descriptor: | Deoxyhypusine synthase, Eukaryotic translation initiation factor 5A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wator, E, Wilk, P, Biela, A.P, Rawski, M, Grudnik, P. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
