7KL5
 
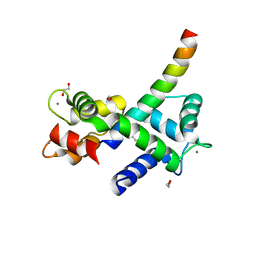 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
2M28
 
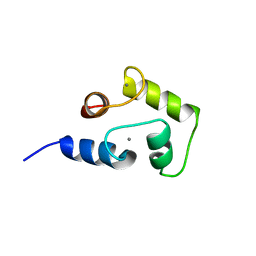 | | NMR structure of Ca2+ bound CaBP4 C-domain | | Descriptor: | CALCIUM ION, Calcium-binding protein 4 | | Authors: | Ames, J.B. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignment of Ca2+ bound CaPB4
To be Published
|
|
2M29
 
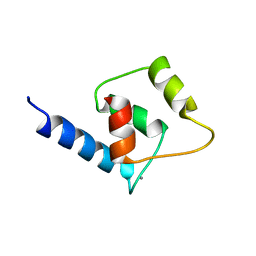 | | NMR structure of Ca2+ bound CaBP4 N-domain | | Descriptor: | CALCIUM ION, Calcium-binding protein 4 | | Authors: | Ames, J.B. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignment of Ca2+ bound CaPB4
To be Published
|
|
3UJ0
 
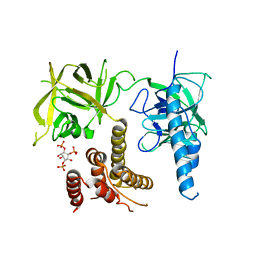 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
3UJ4
 
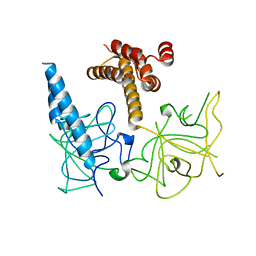 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
6NL3
 
 | | Solution structure of human Coa6 | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog | | Authors: | Naik, M.T, Soma, S, Gohil, V. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase.
Cell Rep, 29, 2019
|
|
2L7Z
 
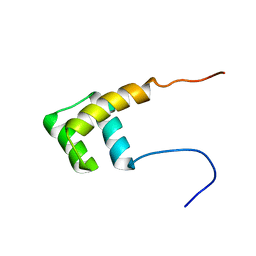 | | NMR Structure of A13 homedomain | | Descriptor: | Homeobox protein Hox-A13 | | Authors: | Ames, J. | | Deposit date: | 2010-12-27 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence specific DNA binding and protein dimerization of HOXA13.
Plos One, 6, 2011
|
|
2LVV
 
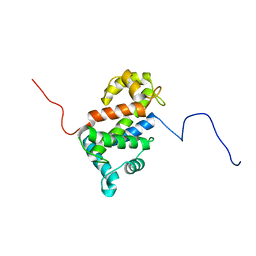 | | NMR structure of TB24 | | Descriptor: | Flagellar calcium-binding protein TB-24 | | Authors: | Ames, J. | | Deposit date: | 2012-07-11 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the calflagin Tb24 flagellar calcium binding protein of Trypanosoma brucei.
Protein Sci., 21, 2012
|
|
2JUL
 
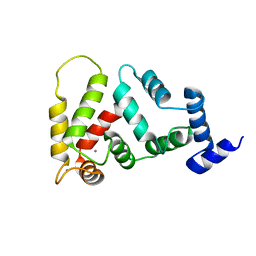 | | NMR Structure of DREAM | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Ames, J. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization.
Biochemistry, 47, 2008
|
|
2JU0
 
 | | Structure of Yeast Frequenin bound to PdtIns 4-kinase | | Descriptor: | CALCIUM ION, Calcium-binding protein NCS-1, Phosphatidylinositol 4-kinase PIK1 | | Authors: | Ames, J. | | Deposit date: | 2007-08-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1).
J.Biol.Chem., 282, 2007
|
|
2KPH
 
 | | NMR Structure of AtraPBP1 at pH 4.5 | | Descriptor: | Pheromone binding protein | | Authors: | Ames, J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Navel Orangeworm Moth Pheromone-Binding Protein (AtraPBP1): Implications for pH-Sensitive Pheromone Detection .
Biochemistry, 49, 2010
|
|
3NB9
 
 | | Crystal structure of radical H88Q Synechocystis sp. PCYA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Gae, D.D. | | Deposit date: | 2010-06-03 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3NB8
 
 | | Crystal structure of oxidized H88Q Synechocystis sp. PCYA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Gae, D.D. | | Deposit date: | 2010-06-03 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3OGN
 
 | |
2LD5
 
 | |
2LLO
 
 | |
2LLQ
 
 | |
3F0M
 
 | | Crystal structure of radical D105N Synechocystis sp. PcyA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Kohler, A.C, Gae, D.D. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
3F0L
 
 | | Crystal structure of oxidized D105N Synechocystis sp. PcyA | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Kohler, A.C, Gae, D.D. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for hydration dynamics in radical stabilization of bilin reductase mutants.
Biochemistry, 49, 2010
|
|
