3QPL
 
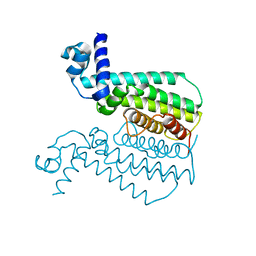 | | G106W mutant of EthR from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Willery, E, Hoos, S, Lecat-Guillet, N, Frenois, F, Dirie, B, Villeret, V, England, P, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-02-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
6H02
 
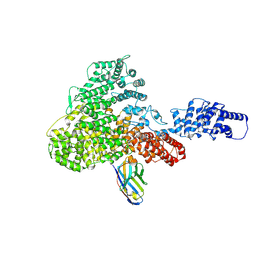 | |
1HZT
 
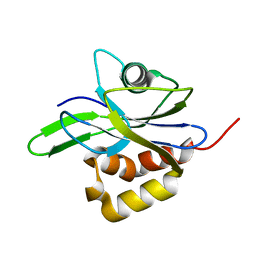 | | CRYSTAL STRUCTURE OF METAL-FREE ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
1HX3
 
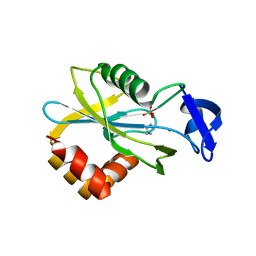 | | CRYSTAL STRUCTURE OF E.COLI ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | IMIDAZOLE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
4RM6
 
 | | Crystal structure of Hemopexin Binding Protein | | Descriptor: | Heme/hemopexin-binding protein | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
4RT6
 
 | | Structure of a complex between hemopexin and hemopexin binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heme/hemopexin-binding protein, Hemopexin | | Authors: | Zambolin, S, Clantin, B, Haouz, A, Villeret, V, Delepelaire, P. | | Deposit date: | 2014-11-13 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for haem piracy from host haemopexin by Haemophilus influenzae.
Nat Commun, 7, 2016
|
|
3SFI
 
 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 5,5,5-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, Transcriptional regulatory repressor protein (TETR-family) | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
3SDG
 
 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 4,4,4-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
4DW6
 
 | | Novel N-phenyl-phenoxyacetamide derivatives as potential EthR inhibitors and ethionamide boosters. Discovery and optimization using High-Throughput Synthesis. | | Descriptor: | AMMONIUM ION, GLYCEROL, HTH-type transcriptional regulator EthR, ... | | Authors: | Flipo, M, Willand, N, Lecat-Guillet, N, Hounsou, C, Desroses, M, Leroux, F, Lens, Z, Villeret, V, Wohlkonig, A, Wintjens, R, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, Baulard, A.R, Deprez, B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel N-phenylphenoxyacetamide derivatives as EthR inhibitors and ethionamide boosters by combining high-throughput screening and synthesis.
J.Med.Chem., 55, 2012
|
|
4FPP
 
 | | Bacterial phosphotransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fioravanti, A, Clantin, B, Dewitte, F, Lens, Z, Verger, A, Biondi, E, Villeret, V. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into ChpT, an essential dimeric histidine phosphotransferase regulating the cell cycle in Caulobacter crescentus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3MPL
 
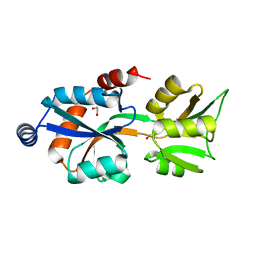 | | Crystal Structure of Bordetella pertussis BvgS VFT2 domain (Double Mutant F375E/Q461E) | | Descriptor: | 1,2-ETHANEDIOL, Virulence sensor protein bvgS | | Authors: | Herrou, J, Bompard, C, Wintjens, R, Dupre, E, Willery, E, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Periplasmic domain of the sensor-kinase BvgS reveals a new paradigm for the Venus flytrap mechanism.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MPK
 
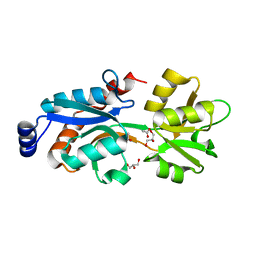 | | Crystal Structure of Bordetella pertussis BvgS periplasmic VFT2 domain | | Descriptor: | ACETATE ION, GLYCEROL, Virulence sensor protein bvgS | | Authors: | Herrou, J, Bompard, C, Wintjens, R, Dupre, E, Willery, E, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Periplasmic domain of the sensor-kinase BvgS reveals a new paradigm for the Venus flytrap mechanism.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2F5X
 
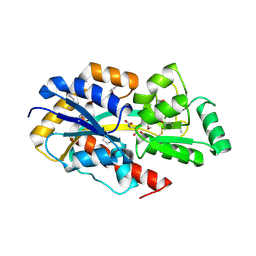 | | Structure of periplasmic binding protein BugD | | Descriptor: | ASPARTIC ACID, BugD | | Authors: | Huvent, I, Belrhali, H, Antoine, R, Bompard, C, Jacob-Dubuisson, F, Villeret, V. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Bordetella pertussis BugD Solute Receptor Unveils the Basis of Ligand Binding in a New Family of Periplasmic Binding Proteins
J.Mol.Biol., 356, 2006
|
|
3NJT
 
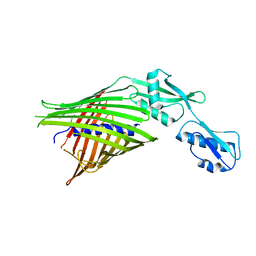 | | Crystal structure of the R450A mutant of the membrane protein FhaC | | Descriptor: | Filamentous hemagglutinin transporter protein fhaC | | Authors: | Clantin, B, Delattre, A.S, Jacob-Dubuisson, F, Villeret, V. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily.
Febs J., 277, 2010
|
|
4Q0C
 
 | | 3.1 A resolution crystal structure of the B. pertussis BvgS periplasmic domain | | Descriptor: | Virulence sensor protein BvgS | | Authors: | Dupre, E, Herrou, J, Lensink, M.F, Wintjens, R, Lebedev, A, Crosson, S, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2014-04-01 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS.
Plos Pathog., 11, 2015
|
|
3TP0
 
 | | Structural activation of the transcriptional repressor EthR from M. tuberculosis by single amino-acid change mimicking natural and synthetic ligands | | Descriptor: | 3-oxo-3-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}propanenitrile, HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Blondiaux, N, Willery, E, Hoos, S, Lecat-Guillet, N, Lens, Z, Wohlkonig, A, Wintjens, R, Soror, S, Fr nois, F, Diri, B, Villeret, V, England, P, Lippens, G, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2011
|
|
7NTT
 
 | |
7NTW
 
 | |
2L23
 
 | |
4I84
 
 | | The crystal structure of the Haemophilus influenzae HxuA secretion domain involved in the two-partner secretion pathway | | Descriptor: | Heme/hemopexin-binding protein | | Authors: | Baelen, S, Dewitte, F, Clantin, B, Villeret, V. | | Deposit date: | 2012-12-03 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the secretion domain of HxuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4M3F
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | HTH-type transcriptional regulator EthR, N-(3-methylbutyl)-4-(2-methyl-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3D
 
 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-{3-[(phenylsulfonyl)amino]prop-1-yn-1-yl}-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3E
 
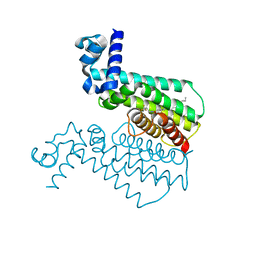 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-{[(propylsulfonyl)amino]methyl}-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
4M3G
 
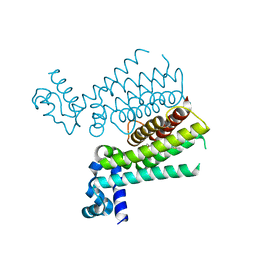 | | Rapid and efficient design of new inhibitors of Mycobacterium tuberculosis transcriptional repressor EthR using fragment growing, merging and linking approaches | | Descriptor: | 4-(2-methyl-1,3-thiazol-4-yl)-N-(3,3,3-trifluoropropyl)benzenesulfonamide, HTH-type transcriptional regulator EthR | | Authors: | Villemagne, B, Flipo, M, Blondiaux, N, Crauste, C, Malaquin, S, Leroux, F, Piveteau, C, Villeret, V, Brodin, P, Villoutreix, B, Sperandio, O, Wohlkonig, A, Wintjens, R, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand Efficiency Driven Design of New Inhibitors of Mycobacterium tuberculosis Transcriptional Repressor EthR Using Fragment Growing, Merging, and Linking Approaches.
J.Med.Chem., 57, 2014
|
|
1RWR
 
 | |
