6S4Q
 
 | | scdSav(SASK) - Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes | | Descriptor: | GLYCEROL, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-06-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Breaking Symmetry: Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes.
J.Am.Chem.Soc., 141, 2019
|
|
6LJR
 
 | | human galectin-16 R55N/H57R | | Descriptor: | Galectin-16, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6S50
 
 | |
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6LBP
 
 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | Descriptor: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | Authors: | Yi, Z, Cao, X, Han, F, Feng, Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
2Y2T
 
 | |
2Y2Y
 
 | | Oxidised form of E. coli CsgC | | Descriptor: | ACETATE ION, CURLI PRODUCTION PROTEIN CSGC | | Authors: | Taylor, J.D, Salgado, P.S, Constable, S.C, Cota, E, Mathews, S.J. | | Deposit date: | 2010-12-16 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic Resolution Insights Into Curli Fiber Biogenesis.
Structure, 19, 2011
|
|
6KXA
 
 | | Galectin-3 CRD binds to GalA dimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
5ZUG
 
 | | Structure of the bacterial acetate channel SatP | | Descriptor: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | Authors: | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
6KXB
 
 | | Galectin-3 CRD binds to GalA trimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
6LOM
 
 | |
6LSB
 
 | |
6BE0
 
 | | AvrA delL154 with IP6, CoA | | Descriptor: | AvrA, COENZYME A, INOSITOL HEXAKISPHOSPHATE | | Authors: | Labriola, J.M, Nagar, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural Analysis of the Bacterial Effector AvrA Identifies a Critical Helix Involved in Substrate Recognition.
Biochemistry, 57, 2018
|
|
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
6BVU
 
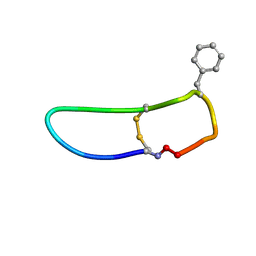 | | SFTI-HFRW-1 | | Descriptor: | Trypsin inhibitor 1 HFRW-1 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6LSD
 
 | |
6BVW
 
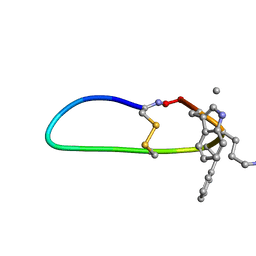 | | SFTI-HFRW-3 | | Descriptor: | Trypsin inhibitor 1 HFRW-3 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-14 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVX
 
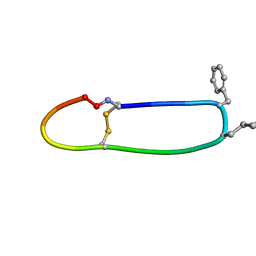 | | SFTI-HFRW-2 | | Descriptor: | Trypsin inhibitor 1 HFRW-2 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-14 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
6BVY
 
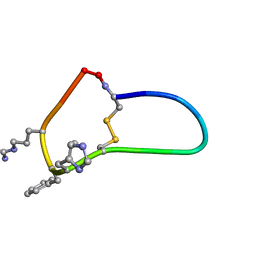 | | SFTI-HFRW-4 | | Descriptor: | Trypsin inhibitor 1 HFRW-4 | | Authors: | Schroeder, C.I, White, A. | | Deposit date: | 2017-12-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
6LS6
 
 | |
7WD0
 
 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
 | | RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
