5T27
 
 | | mPI3Kd IN COMPLEX WITH 5d | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-[(1~{S})-1-morpholin-4-ylethyl]-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T23
 
 | | PI3Kg IN COMPLEX WITH 5d | | Descriptor: | 3-(benzotriazol-1-yl)-5-[1-methyl-5-[(1~{S})-1-morpholin-4-ylethyl]-1,2,4-triazol-3-yl]pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2B
 
 | | mPI3Kd IN COMPLEX WITH 5e | | Descriptor: | 5-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]-3-(1~{H}-pyrazol-4-yl)pyrazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2I
 
 | | mPI3Kd IN COMPLEX WITH 7k | | Descriptor: | 1-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]pyrazole-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2D
 
 | | mPI3Kd IN COMPLEX WITH 7j | | Descriptor: | 4-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]-~{N},~{N}-dimethyl-pyrazole-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T2M
 
 | | mPI3Kd IN COMPLEX WITH 7m | | Descriptor: | 4-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4GLJ
 
 | | Crystal structure of methylthioadenosine phosphorylase in complex with rhodamine B | | Descriptor: | CHLORIDE ION, N-[9-(2-carboxyphenyl)-6-(diethylamino)-3H-xanthen-3-ylidene]-N-ethylethanaminium, PHOSPHATE ION, ... | | Authors: | Bujacz, A, Bujacz, G, Cieslinski, H, Bartasun, P. | | Deposit date: | 2012-08-14 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A study on the interaction of rhodamine B with methylthioadenosine phosphorylase protein sourced from an antarctic soil metagenomic library.
Plos One, 8, 2013
|
|
5T2G
 
 | | mPI3Kd IN COMPLEX WITH 7i | | Descriptor: | 4-[3-azanyl-6-[1-methyl-5-(1-phenylcyclopropyl)-1,2,4-triazol-3-yl]pyrazin-2-yl]pyrazole-1-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J, Terstige, I, Perry, M, Svensson, T, Tyrchan, C, Lindmark, H, Oster, L. | | Deposit date: | 2016-08-23 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of triazole aminopyrazines as a highly potent and selective series of PI3K delta inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1JLJ
 
 | | 1.6 Angstrom crystal structure of the human neuroreceptor anchoring and molybdenum cofactor biosynthesis protein gephyrin | | Descriptor: | FORMIC ACID, SODIUM ION, gephyrin | | Authors: | Schwarz, G, Schrader, N, Mendel, R.R, Hecht, H.-J, Schindelin, H. | | Deposit date: | 2001-07-16 | | Release date: | 2001-09-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human gephyrin and plant Cnx1 G domains: comparative analysis and functional implications.
J.Mol.Biol., 312, 2001
|
|
3ESW
 
 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
1JW9
 
 | | Structure of the Native MoeB-MoaD Protein Complex | | Descriptor: | MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, SUBUNIT 1, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
2B3F
 
 | | Thermus thermophilus Glucose/Galactose Binding Protein Bound With Galactose | | Descriptor: | beta-D-galactopyranose, glucose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Warren, J.J, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2005-09-20 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The crystal structure of a thermophilic glucose binding protein reveals adaptations that interconvert mono and di-saccharide binding sites.
J.Mol.Biol., 362, 2006
|
|
1JWA
 
 | | Structure of the ATP-bound MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
1JIZ
 
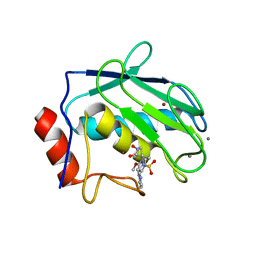 | | Crystal Structure Analysis of human Macrophage Elastase MMP-12 | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, ZINC ION, ... | | Authors: | Nar, H, Werle, K, Bauer, M.M.T, Dollinger, H, Jung, B. | | Deposit date: | 2001-07-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human macrophage elastase (MMP-12) in complex with a hydroxamic acid inhibitor.
J.Mol.Biol., 312, 2001
|
|
1JWB
 
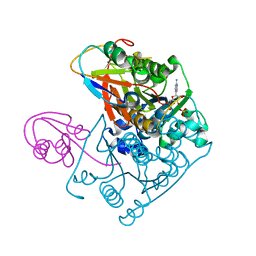 | | Structure of the Covalent Acyl-Adenylate Form of the MoeB-MoaD Protein Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MOLYBDOPTERIN BIOSYNTHESIS MOEB PROTEIN, MOLYBDOPTERIN [MPT] CONVERTING FACTOR, ... | | Authors: | Lake, M.W, Wuebbens, M.M, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2001-09-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex.
Nature, 414, 2001
|
|
4ABM
 
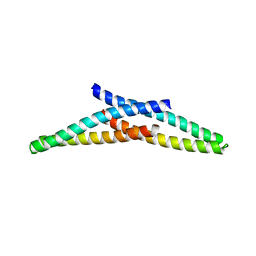 | | Crystal Structure of CHMP4B hairpin | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 4B | | Authors: | Martinelli, N, Hartlieb, B, Usami, Y, Sabin, C, Dordor, A, Ribeiro, E.A, Gottlinger, H, Weissenhorn, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cc2D1A is a Regulator of Escrt-III Chmp4B.
J.Mol.Biol., 419, 2012
|
|
4BKM
 
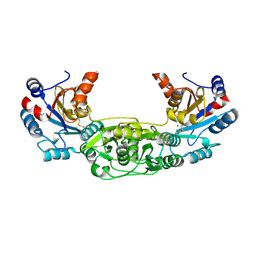 | | Crystal structure of the murine AUM (phosphoglycolate phosphatase) capping domain as a fusion protein with the catalytic core domain of murine chronophin (pyridoxal phosphate phosphatase) | | Descriptor: | MAGNESIUM ION, NITRATE ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Seifried, A, Gohla, A, Schindelin, H. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evolutionary and Structural Analyses of the Mammalian Haloacid Dehalogenase-Type Phosphatases Aum and Chronophin Provide Insight Into the Basis of Their Different Substrate Specificities.
J.Biol.Chem., 289, 2014
|
|
2NQN
 
 | | MoeA T100W | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQR
 
 | | MoeA D142N | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
4BX3
 
 | |
2NQV
 
 | | MoeA D228A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nciolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
4BX2
 
 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in complex with Beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop.
J.Biol.Chem., 289, 2014
|
|
2NQS
 
 | | MoeA E188A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQU
 
 | | MoeA E188Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQQ
 
 | | MoeA R137Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
