7ZN5
 
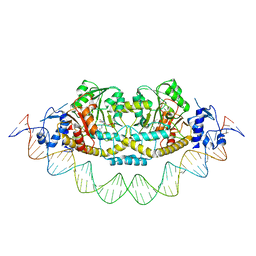 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
2DSM
 
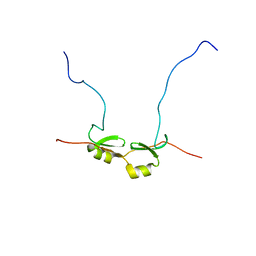 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
2E4G
 
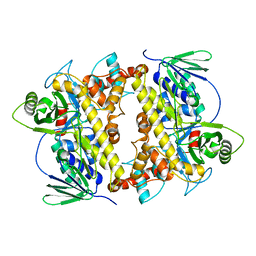 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
2BJM
 
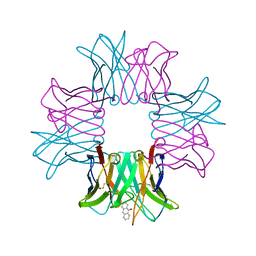 | | SPE7:Anthrone Complex | | Descriptor: | ANTHRONE, IGE SPE7 HEAVY CHAIN, IGE SPE7 LIGHT CHAIN | | Authors: | James, L.C, Tawfik, D.S. | | Deposit date: | 2005-02-04 | | Release date: | 2005-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Kinetics of a Transient Antibody Binding Intermediate Reveal a Kinetic Discrimination Mechanism in Antigen Recognition
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C8V
 
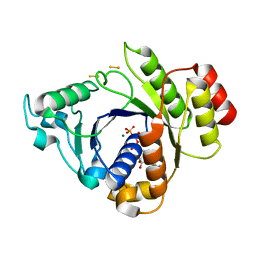 | | Insights into the role of nucleotide-dependent conformational change in nitrogenase catalysis: Structural characterization of the nitrogenase Fe protein Leu127 deletion variant with bound MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Sen, S, Krishnakumar, A, McClead, J, Johnson, M.K, Seefeldt, L.C, Szilagyi, R.K, Peters, J.W. | | Deposit date: | 2005-12-08 | | Release date: | 2006-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Role of Nucleotide-Dependent Conformational Change in Nitrogenase Catalysis: Structural Characterization of the Nitrogenase Fe Protein Leu127 Deletion Variant with Bound Mgatp.
J.Inorg.Biochem., 100, 2006
|
|
2FJ6
 
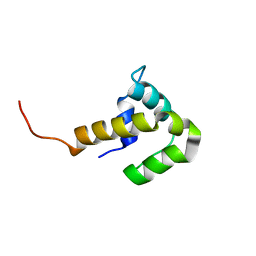 | | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391. | | Descriptor: | Hypothetical UPF0346 protein yozE | | Authors: | Rossi, P, Acton, T.B, Cunningham, K.E, Ma, L.C, Shetty, K, Swapna, G.V.T, Xiao, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0346 protein yozE from Bacillus subtilis. Northeast Structural Genomics target SR391.
To be Published
|
|
2EST
 
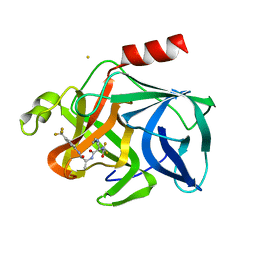 | |
2FCU
 
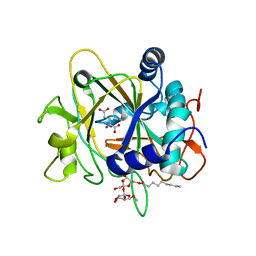 | | SyrB2 with alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, syringomycin biosynthesis enzyme 2 | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FCT
 
 | | SyrB2 with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FMS
 
 | | DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FMQ
 
 | | Sodium in active site of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FVT
 
 | | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris: Northeast Structural Genomics Target RpR43 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Ho, C.K, Cunningham, K, Ma, L.C, Conover, K, Xiao, R, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Rpa2829 protein from Rhodopseudomonas palustris
To be Published
|
|
2FMP
 
 | | DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Batra, V.K, Beard, W.A, Shock, D.D, Krahn, J.M, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Magnesium-induced assembly of a complete DNA polymerase catalytic complex.
Structure, 14, 2006
|
|
2FCV
 
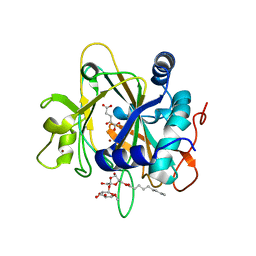 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
2FFG
 
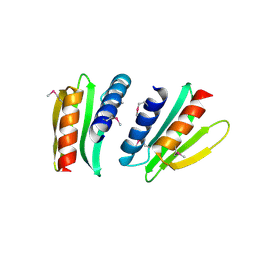 | | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360. | | Descriptor: | ykuJ | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Ho, C.K, Janjua, H, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360.
To be Published
|
|
2H30
 
 | | Crystal structure of the N-terminal domain of PilB from Neisseria gonorrhoeae | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Brot, N, Collet, J.F, Johnson, L.C, Jonsson, T.J, Weissbach, H, Lowther, W.T. | | Deposit date: | 2006-05-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Thioredoxin Domain of Neisseria gonorrhoeae PilB Can Use Electrons from DsbD to Reduce Downstream Methionine Sulfoxide Reductases.
J.Biol.Chem., 281, 2006
|
|
2GZO
 
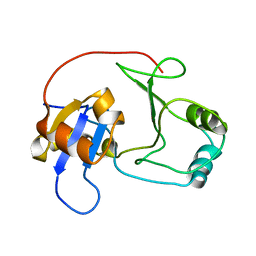 | | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39 | | Descriptor: | UPF0301 protein SO3346 | | Authors: | Singarapu, K.K, Liu, G, Eletsky, A, Xu, D, Sukumaran, D.K, Mei, J, Xiao, R, Cunningham, K, Ma, L.C, Ritu, S, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39
To be Published
|
|
4GBI
 
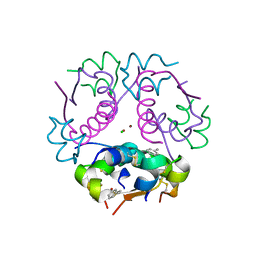 | | Crystal structure of aspart insulin at pH 6.5 | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Lima, L.M.T.R, Favero-Retto, M.P, Palmieri, L.C. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | A T3R3 hexamer of the human insulin variant B28Asp.
Biophys.Chem., 173, 2013
|
|
4GBL
 
 | | Crystal structure of aspart insulin at pH 8.5 | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Lima, L.M.T.R, Favero-Retto, M.P, Palmieri, L.C. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A T3R3 hexamer of the human insulin variant B28Asp.
Biophys.Chem., 173, 2013
|
|
3OB4
 
 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
3OTS
 
 | | MDR769 HIV-1 protease complexed with MA/CA hepta-peptide | | Descriptor: | MA/CA substrate peptide, Multi-drug resistant HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OTY
 
 | | MDR769 HIV-1 protease complexed with RT/RH hepta-peptide | | Descriptor: | MDR HIV-1 protease, RT/RH substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUA
 
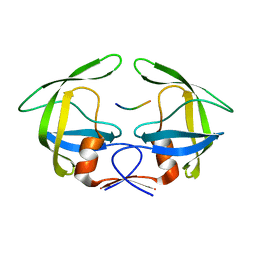 | | MDR769 HIV-1 protease complexed with p1/p6 hepta-peptide | | Descriptor: | HIV-1 protease, p1/p6 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OWV
 
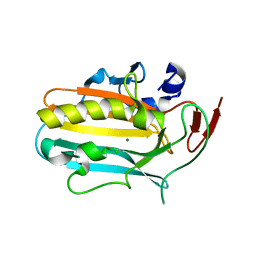 | | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DNA-entry nuclease, MAGNESIUM ION | | Authors: | Moon, A.F, Midon, M, Meiss, G, Pingoud, A.M, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into catalytic and substrate binding mechanisms of the strategic EndA nuclease from Streptococcus pneumoniae.
Nucleic Acids Res., 39, 2011
|
|
