1MNH
 
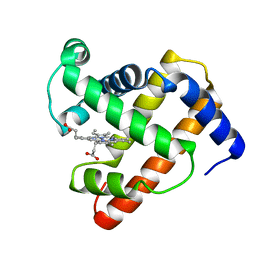 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Davies, G.J, Wilkinson, A.J. | | Deposit date: | 1995-01-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1K9T
 
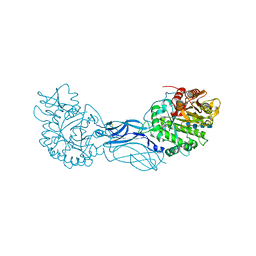 | | Chitinase a complexed with tetra-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Prag, G, Tucker, P.A, Oppenheim, A.B. | | Deposit date: | 2001-10-30 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
1LT3
 
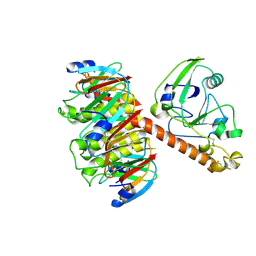 | | HEAT-LABILE ENTEROTOXIN DOUBLE MUTANT N40C/G166C | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-12 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit.
Protein Sci., 6, 1997
|
|
1LT6
 
 | |
1LTT
 
 | |
1LT4
 
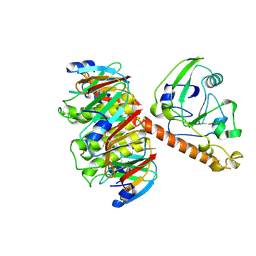 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
1KV6
 
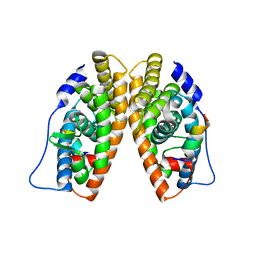 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
1NAW
 
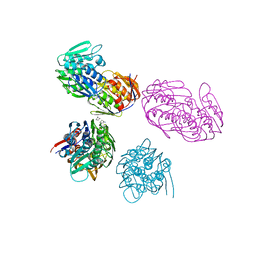 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
1PBL
 
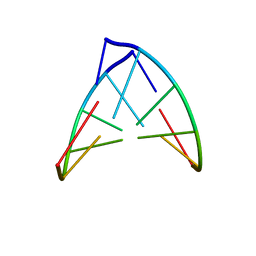 | | STRUCTURE OF RIBONUCLEIC ACID, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*OMCP*OMGP*OMCP*OMGP*OMCP*OMG)-3') | | Authors: | Popenda, M, Biala, E, Milecki, J, Adamiak, R.W. | | Deposit date: | 1996-08-05 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2'-O-Me(CGCGCG)2 under low salt conditions.
Nucleic Acids Res., 25, 1997
|
|
1NLU
 
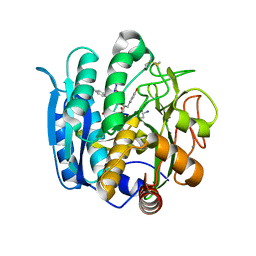 | | Pseudomonas sedolisin (serine-carboxyl proteinase) complexed with two molecules of pseudo-iodotyrostatin | | Descriptor: | CALCIUM ION, PSEUDO-IODOTYROSTATIN, SEDOLISIN | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Glodfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Two inhibitor molecules bound in the active site of Pseudomonas sedolisin: a model for the bi-product complex following cleavage of a peptide substrate.
Biochem.Biophys.Res.Commun., 314, 2004
|
|
1QAW
 
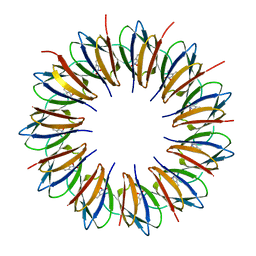 | | Regulatory Features of the TRP Operon and the Crystal Structure of the TRP RNA-Binding Attenuation Protein from Bacillus Stearothermophilus. | | Descriptor: | TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Chen, X.-P, Antson, A.A, Yang, M, Baumann, C, Dodson, E.J, Dodson, G.G, Gollnick, P. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J.Mol.Biol., 289, 1999
|
|
1QN0
 
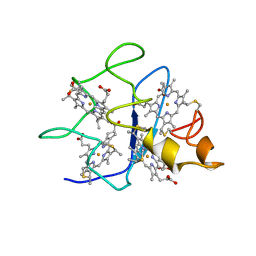 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1QN1
 
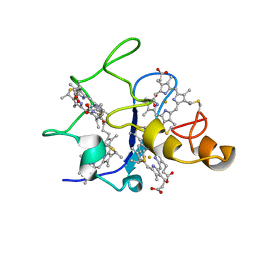 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1PZB
 
 | |
1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1QKN
 
 | |
1QKM
 
 | |
1PZA
 
 | |
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1GWQ
 
 | |
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HEU
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GM4
 
 | | OXIDISED STRUCTURE OF CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-10 | | Release date: | 2002-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome.
J.Biol.Chem., 276, 2001
|
|
1GMB
 
 | | Reduced structure of CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 at pH 7.6 | | Descriptor: | CYTOCHROME C3, HEME C, SULFATE ION | | Authors: | Bento, I, Louro, R, Matias, P.M, Catarino, T, Baptista, A.M, Soares, C.M, Carrondo, M.A, Turner, D.L, Xavier, A.V. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Component in the Coupled Transfer of Multiple Electrons and Protons in a Monomeric Tetraheme Cytochrome
J.Biol.Chem., 276, 2001
|
|
