7NLN
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLS
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with methyl 4-hydroxy-3-iodobenzoate | | Descriptor: | Acetylglutamate kinase, SULFATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLQ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(isoxazol-5-yl)phenol | | Descriptor: | 2-(1,2-oxazol-5-yl)phenol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLP
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-canavanine | | Descriptor: | Acetylglutamate kinase, L-CANAVANINE, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLO
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-arginine | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ARGININE, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(5-methoxy-1H-indol-3-yl)acetonitrile | | Descriptor: | 2-(5-methoxy-1~{H}-indol-3-yl)ethanenitrile, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in apo form. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
5LI6
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
3DDI
 
 | | Crystal structure of the mimivirus NDK +Kpn-N62L-R107G triple mutant complexed with TDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, A. | | Deposit date: | 2008-06-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
7NNQ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with nicotinamide adenine dinucleotide phosphate (NADP+) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-gamma-glutamyl-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NPH
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 5-methoxy-1,3-benzoxazole-2-carboxylic acid | | Descriptor: | 5-methoxy-1,3-benzoxazole-2-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase, PHOSPHATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLX
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 7-(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 7-(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN4
 
 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate and 3-hydroxy-2-naphthoic acid. | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 5-Methoxy-6-(trifluoromethyl)indole. | | Descriptor: | 5-methoxy-6-(trifluoromethyl)-1~{H}-indole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNC
 
 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal-5'-phosphate and 6-Methoxyquinoline-3-carboxylic acid | | Descriptor: | 6-methoxyquinoline-3-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with xanthene-9-carboxylic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9~{H}-xanthene-9-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NM0
 
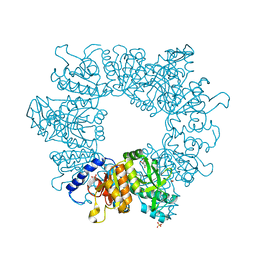 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNI
 
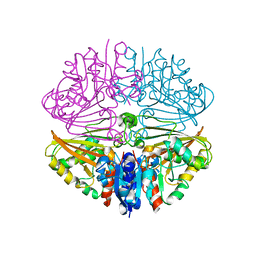 | |
7NLY
 
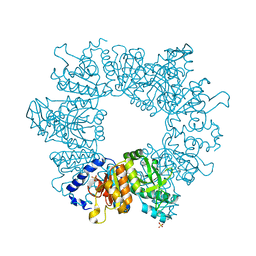 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-Chlorobenzimidazole. | | Descriptor: | 2-chloranyl-1~{H}-benzimidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNB
 
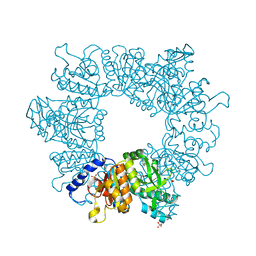 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2,8-bis(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 1,2-ETHANEDIOL, 2,8-bis(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN8
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1H-indole-3-carbonitrile. | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indole-3-carbonitrile, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN1
 
 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate | | Descriptor: | Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NPJ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 6-phenoxy-3-pyridinamine | | Descriptor: | 6-phenoxy-3-pyridinamine, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-27 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NOT
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with nicotinamide adenine dinucleotide phosphate (NADP+) and 5-Methoxy-3-indoleacetic acid | | Descriptor: | (5-methoxy-1H-indol-3-yl)acetic acid, N-acetyl-gamma-glutamyl-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
