4MH7
 
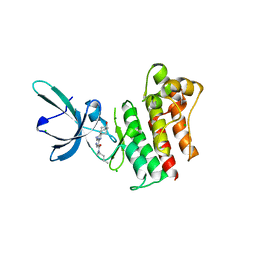 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1896 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-butyl-2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-methylpyrimidine-5-carboxamide, ... | | Authors: | Zhang, W, McIver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, M.J, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, DiPaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
3FVR
 
 | |
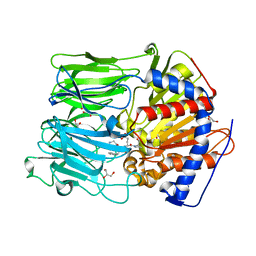
1UOP
 
 | |
3FYT
 
 | |
2H8W
 
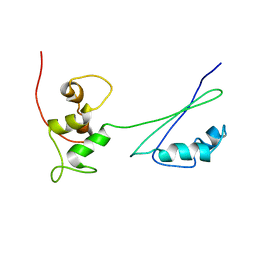 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
1ZMX
 
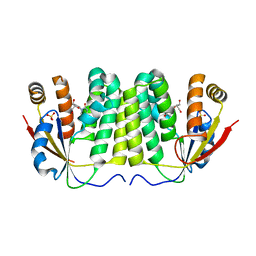 | | Crystal structure of D. melanogaster deoxyribonucleoside kinase N64D mutant in complex with thymidine | | Descriptor: | Deoxynucleoside kinase, SULFATE ION, THYMIDINE | | Authors: | Welin, M, Skovgaard, T, Knecht, W, Berenstein, D, Munch-Petersen, B, Piskur, J, Eklund, H. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the changed substrate specificity of Drosophila melanogaster deoxyribonucleoside kinase mutant N64D.
Febs J., 272, 2005
|
|
1UOO
 
 | |
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
1ZJY
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
4RRN
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'S,10a'R)-2-amino-8'-(2-fluoropyridin-3-yl)-1-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
3ZNX
 
 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZJG
 
 | | A20 OTU domain with irreversibly oxidised Cys103 from 60 min H2O2 soak. | | Descriptor: | CHLORIDE ION, TUMOR NECROSIS FACTOR ALPHA-INDUCED PROTEIN 3 | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
6EHO
 
 | | Dimer of the Sortilin Vps10p domain at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thirup, S.S, Quistgaard, E.H, Januliene, D, Andersen, J.L, Nielsen, J.A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Acidic Environment Induces Dimerization and Ligand Binding Site Collapse in the Vps10p Domain of Sortilin.
Structure, 25, 2017
|
|
1FDW
 
 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 MUTANT H221Q COMPLEXED WITH ESTRADIOL | | Descriptor: | 17-BETA-HYDROXYSTEROID DEHYDROGENASE, ESTRADIOL | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unusual charge stabilization of NADP+ in 17beta-hydroxysteroid dehydrogenase.
J.Biol.Chem., 273, 1998
|
|
6TAZ
 
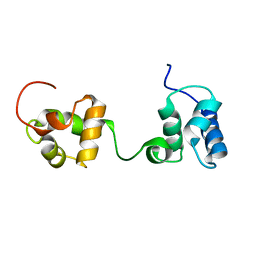 | | Timeless couples G quadruplex detection with processing by DDX11 during DNA replication | | Descriptor: | Protein timeless homolog | | Authors: | Lerner Koch, L, Holzer, S, Kilkenny, M.L, Murat, P, Svikovic, S, Schiavone, D, Bittleston, A, Maman, J.D, Branzei, D, Stott, K, Pellegrini, L, Sale, E.J. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication.
Embo J., 39, 2020
|
|
6EIE
 
 | |
8AH0
 
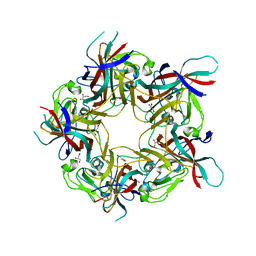 | | BK Polyomavirus VP1 mutant VQQ | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
8AH1
 
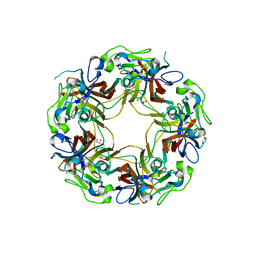 | | BK Polyomavirus VP1 mutant N-Q | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1 | | Authors: | Sorin, M.N, Di Maio, A, Silva, L.M, Ebert, D, Delannoy, C, Nguyen, N.-K, Guerardel, Y, Chai, W, Halary, F, Renaudin-Autain, K, Liu, Y, Bressollette-Bodin, C, Stehle, T, McIlroy, D. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural and functional analysis of natural capsid variants suggests sialic acid-independent entry of BK polyomavirus.
Cell Rep, 42, 2023
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
4PSL
 
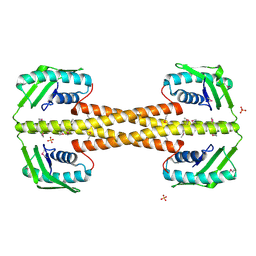 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form I) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Sohmen, D, Wilson, D.N, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
1ZK0
 
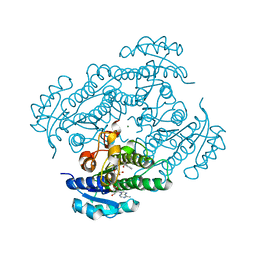 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
6TLZ
 
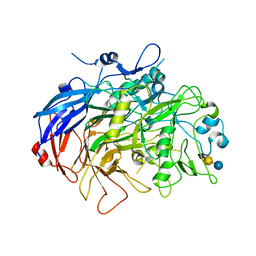 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 3'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
4A0X
 
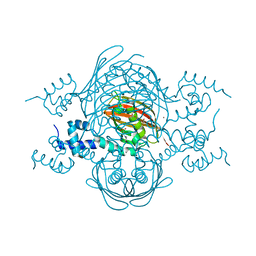 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, TRANSCRIPTION FACTOR FAPR, ... | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
