8A8N
 
 | |
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
8ASY
 
 | | SARS-CoV-2 Omicron BA.2.75 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-08-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75.
Cell Rep, 42, 2022
|
|
1BND
 
 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
1D4V
 
 | | Crystal structure of trail-DR5 complex | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Mongkolsapaya, J, Grimes, J.M, Stuart, D.I, Jones, E.Y, Screaton, G.R. | | Deposit date: | 1999-10-06 | | Release date: | 1999-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation
Nat.Struct.Biol., 6, 1999
|
|
1E27
 
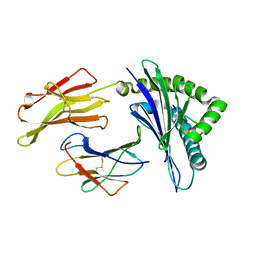 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM1(LPPVVAKEI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HIV-1 PEPTIDE (LPPVVAKEI), HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
1E28
 
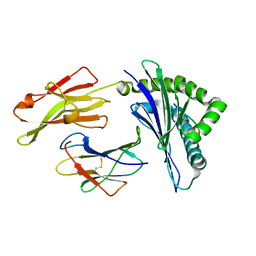 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM2(TAFTIPSI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN, PEPTIDE | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
1FNH
 
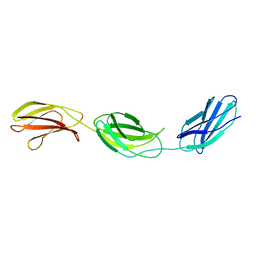 | | CRYSTAL STRUCTURE OF HEPARIN AND INTEGRIN BINDING SEGMENT OF HUMAN FIBRONECTIN | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Sharma, A, Askari, J, Humphries, M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 1999-01-28 | | Release date: | 1999-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a heparin- and integrin-binding segment of human fibronectin.
EMBO J., 18, 1999
|
|
8PAG
 
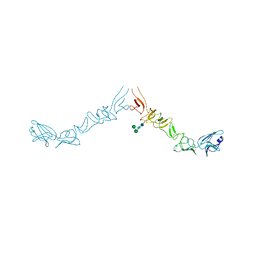 | | Crystal structure of the ectodomain of Norway rat pestivirus E2 glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Aitkenhead, H, Stuart, D.I, EL Omari, K. | | Deposit date: | 2023-06-07 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural comparison of typical and atypical E2 pestivirus glycoproteins.
Structure, 32, 2024
|
|
8PAE
 
 | |
8PAB
 
 | |
3VBF
 
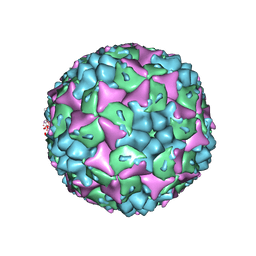 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VBO
 
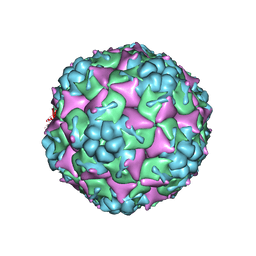 | | Crystal structure of formaldehyde treated empty human Enterovirus 71 particle (cryo at 100K) | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ZN5
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-virus-derived) | | Descriptor: | CHLORIDE ION, VP16 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
4AB6
 
 | | Regulatory domain structure of NMB2055 (MetR), C103S C106S mutant, a LysR family regulator from N. meningitidis | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | Authors: | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4A8Y
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*TP*TP*CP*GP*CP*GP*TP*AP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
4A8W
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*CP*GP*CP*GP*TP*AP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
4A8M
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*AP*AP*TP*CP)-3', ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
4A8S
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*TP*TP*CP*GP*CP*GP*TP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
3ZN6
 
 | | VP16-VP17 complex, a complex of the two major capsid proteins of bacteriophage P23-77 | | Descriptor: | CHLORIDE ION, SODIUM ION, VP16, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3ZN4
 
 | | VP16, a capsid protein of bacteriophage P23-77 (VP16-type-2) | | Descriptor: | CHLORIDE ION, CITRIC ACID, VP16 | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
4A8Q
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*DTP*TP*CP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
4A8F
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*DAP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
