3TYS
 
 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3TYR
 
 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
3TZC
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(Y155F) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
5IJE
 
 | | Crystal structure of Equine Serum Albumin in the presence of 30 mM zinc at pH 7.4 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Szlachta, K, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UDU
 
 | | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, CHLORIDE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Grimshaw, S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
To be Published
|
|
5W8W
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
4MOU
 
 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | Descriptor: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
3GHD
 
 | | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Descriptor: | a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Authors: | Dong, A, Xu, X, Chruszcz, M, Brown, G, Proudfoot, M, Edwards, A.M, Joachimiak, A, Minor, W, Savchenko, A, Yaleunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain
To be Published
|
|
5IJ5
 
 | | Crystal structure of Equine Serum Albumin in the presence of 50 mM zinc at pH 4.5 | | Descriptor: | GLYCEROL, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Majorek, K.A, Shabalin, I.G, Cymborowski, M.T, Zheng, H, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
5IIX
 
 | | Crystal structure of Equine Serum Albumin in the presence of 15 mM zinc at pH 6.5 | | Descriptor: | SULFATE ION, Serum albumin, UNKNOWN LIGAND, ... | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Gasiorowska, O.A, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
2NAR
 
 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
5UOG
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
2MSX
 
 | | The solution structure of the MANEC-type domain from Hepatocyte Growth Factor Inhibitor 1 reveals an unexpected PAN/apple domain-type fold | | Descriptor: | Kunitz-type protease inhibitor 1 | | Authors: | Hong, Z, Nowakowski, M.E, Spronk, C, Petersen, S.V, Petersen, J.S, Kozminski, W, Mulder, F, Jensen, J.K. | | Deposit date: | 2014-08-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold.
Biochem.J., 466, 2015
|
|
3V48
 
 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | Descriptor: | Aminoacrylate peracid reductase RutC | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3HHL
 
 | | Crystal structure of methylated RPA0582 protein | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sledz, P, Niedzialkowska, E, Chruszcz, M, Porebski, P, Yim, V, Kudritska, M, Zimmerman, M.D, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of methylated RPA0582 protein
To be Published
|
|
4PMJ
 
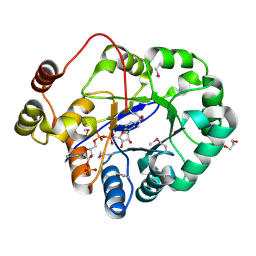 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
3TL2
 
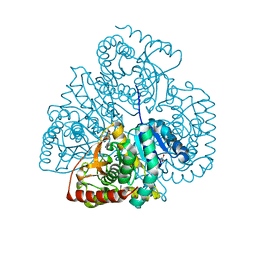 | | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation. | | Descriptor: | 1,2-ETHANEDIOL, Malate dehydrogenase, THIOCYANATE ION | | Authors: | Blus, B.J, Chruszcz, M, Tkaczuk, K.L, Osinski, T, Cymborowski, M, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus anthracis str. Ames malate dehydrogenase in closed conformation.
TO BE PUBLISHED
|
|
4N7C
 
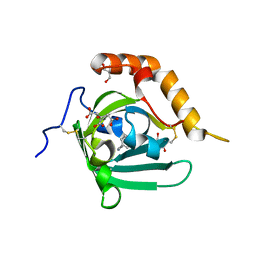 | | Structural re-examination of native Bla g 4 | | Descriptor: | 4-(2-aminoethyl)phenol, Bla g 4 allergen variant 1, CITRIC ACID, ... | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
3UEC
 
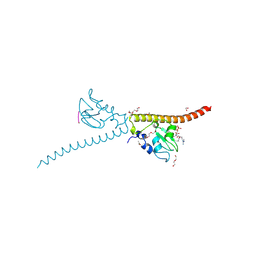 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
4OAE
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol | | Descriptor: | 1,2-ETHANEDIOL, CHLORAMPHENICOL, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of a GNAT superfamily acetyltransferase PA4794 C29A/C117A/Y128A mutant in complex with chloramphenicol
To be Published
|
|
