6T7Y
 
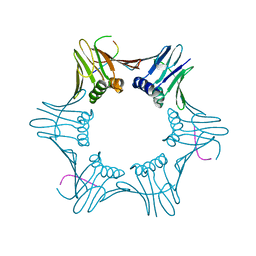 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | Descriptor: | photoswitching chromogenic protein | | Authors: | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
2M5J
 
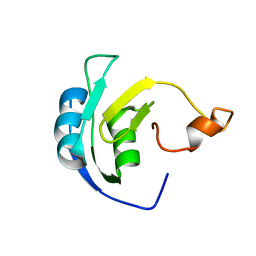 | | Solution structure of the periplasmic signaling domain of HasR, a TonB-dependent outer membrane heme transporter | | Descriptor: | HasR protein | | Authors: | Malki, I, Cardoso de Amorim, G, Prochnicka-Chalufour, A, Simenel, C, Delepierre, M, Izadi-Pruneyre, N. | | Deposit date: | 2013-02-26 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interaction of a Partially Disordered Antisigma Factor with Its Partner, the Signaling Domain of the TonB-Dependent Transporter HasR
Plos One, 9, 2014
|
|
6RQX
 
 | |
8CPE
 
 | |
2NQ3
 
 | | Crystal structure of the C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase | | Descriptor: | CHLORIDE ION, Itchy homolog E3 ubiquitin protein ligase | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase
To be Published
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6IBQ
 
 | | Structure of a nonameric RNA duplex at room temperature in ChipX microfluidic device | | Descriptor: | DNA/RNA (5'-R(*CP*GP*UP*GP*AP*UP*CP*G)-D(P*C)-3'), SULFATE ION | | Authors: | de Wijn, R, Olieric, V, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6T7X
 
 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6IBP
 
 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6HW1
 
 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6RYF
 
 | | High-resolution crystal structure of ERAP1 in complex with 15mer phosphinic peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1.
Proc.Natl.Acad.Sci.USA, 2019
|
|
2OXE
 
 | | Structure of the Human Pancreatic Lipase-related Protein 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic lipase-related protein 2, ... | | Authors: | Walker, J.R, Davis, T, Seitova, A, Finerty Jr, P.J, Butler-Cole, C, Kozieradzki, I, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
2JJJ
 
 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
2JJI
 
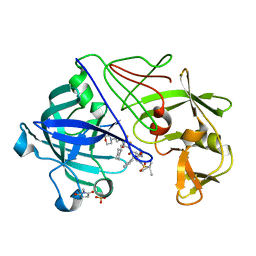 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
2QU6
 
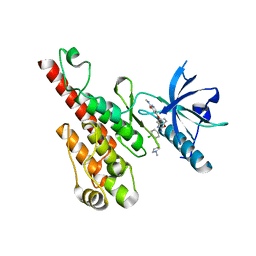 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzoxazole inhibitor | | Descriptor: | 4-({2-[(4-chloro-3-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}phenyl)amino]-1,3-benzoxazol-5-yl}oxy)-N-methylpyridine-2-carboxamide, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
2QU5
 
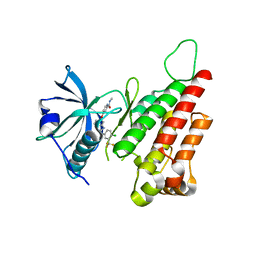 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzimidazole inhibitor | | Descriptor: | 4-[[2-[[4-chloro-3-(trifluoromethyl)phenyl]amino]-3H-benzimidazol-5-yl]oxy]-N-methyl-pyridine-2-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
3IAX
 
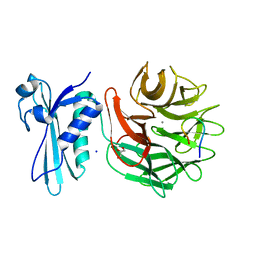 | |
6ZZ9
 
 | |
5TJT
 
 | |
7AXC
 
 | | Crystal structure of the hPXR-LBD in complex with ferutinine | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Nuclear receptor subfamily 1 group I member 2, ... | | Authors: | Granell, M, Delfosse, V, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AXB
 
 | | Crystal structure of the hPXR-LBD in complex with endosulfan | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Nuclear receptor subfamily 1 group I member 2, ... | | Authors: | Huet, T, Delfosse, V, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
