8WGT
 
 | | Crystal structure of V30M-TTR in complex with compound 7 | | Descriptor: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGS
 
 | | Crystal structure of V30M-TTR in complex with compound 4 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-(2-ethyl-4-iodanyl-1-benzofuran-3-yl)methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGU
 
 | | Crystal structure of V30M-TTR in complex with compound 20 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-[2-ethyl-4,7-bis(fluoranyl)-1-benzofuran-3-yl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
5B71
 
 | | Crystal structure of complement C5 in complex with SKY59 | | Descriptor: | Complement C5 beta chain, SKY59 Fab heavy chain, SKY59 Fab light chain | | Authors: | Irie, M, Shimizu, Y, Sampei, Z, Fukuzawa, T. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Long lasting neutralization of C5 by SKY59, a novel recycling antibody, is a potential therapy for complement-mediated diseases.
Sci Rep, 7, 2017
|
|
4ENY
 
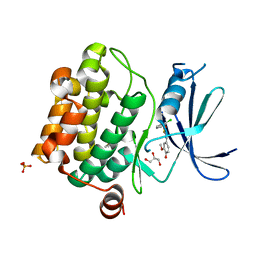 | | Crystal Structure of Pim-1 kinase in complex with (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-methoxybenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-methoxybenzylidene)-1,3-thiazolidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7F4X
 
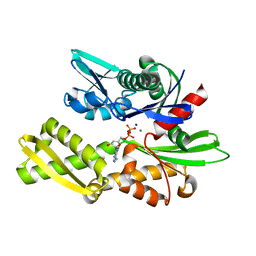 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7Y6L
 
 | |
7Y6N
 
 | |
9JIR
 
 | |
9JIS
 
 | |
9JIQ
 
 | |
4ENX
 
 | | Crystal Structure of Pim-1 Kinase in complex with inhibitor (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-nitrobenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-nitrobenzylidene)-1,3-thiazolidin-4-one, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5EVY
 
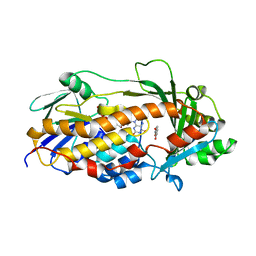 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5ETY
 
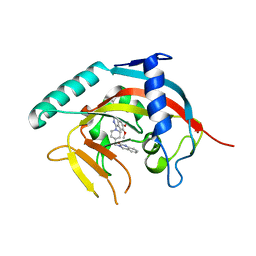 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | Descriptor: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | Authors: | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
6JTB
 
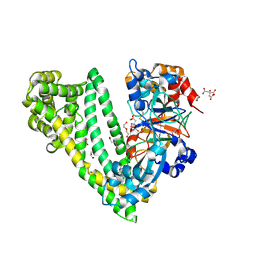 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
8I1C
 
 | |
8I1A
 
 | |
8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1F
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I8S
 
 | |
8I1D
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
