8XKH
 
 | | Structure of chimeric RyR Complex with tetraniliprole | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XLH
 
 | | Structure of chimeric RyR-I4657M/G4819E | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-26 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8XJI
 
 | | Structure of chimeric RyR complex with flubendiamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-21 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8HEH
 
 | | Crystal structure of GCN5-related N-acetyltransferase 05790 | | Descriptor: | COENZYME A, GLYCEROL, GNAT family N-acetyltransferase | | Authors: | Xu, M.X, Ran, T.T, Wang, W. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of prodigiosin binding protein PgbP, a GNAT family protein, in Serratia marcescens FS14.
Biochem.Biophys.Res.Commun., 640, 2022
|
|
8H0M
 
 | | Crystal structure of VioD | | Descriptor: | (2S)-2-ethylhexan-1-ol, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Xu, M, Ran, T, Wang, W. | | Deposit date: | 2022-09-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of the key enzyme VioD in the violacein synthesis pathway.
Proteins, 91, 2023
|
|
5GXT
 
 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
5DPQ
 
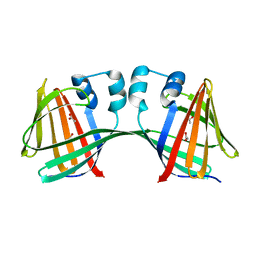 | | Crystal Structure of E72A mutant of domain swapped dimer Human Cellular Retinol Binding Protein | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Geiger, J.H, Borhan, B. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
8XLF
 
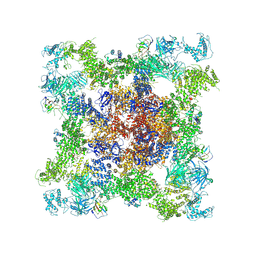 | | Structure of chimeric RyR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Lin, L, Wang, C, Wang, W, Jiang, H, Yuchi, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of ryanodine receptors and diamide insecticides reveal the mechanisms of selectivity and resistance.
Nat Commun, 15, 2024
|
|
8H4E
 
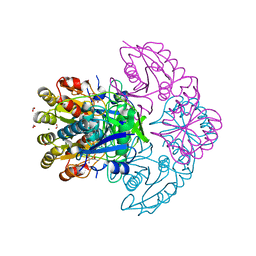 | | Blasnase-T13A/P55N with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H46
 
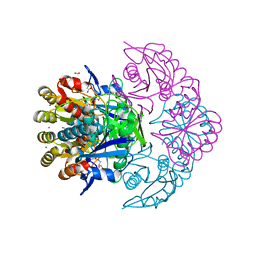 | | Blasnase-T13A/P55N with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, GLYCEROL, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H47
 
 | | Blasnase-T13A/P55F | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4B
 
 | | Blasnase-T13A/M57P with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4D
 
 | | Blasnase-T13A/M57N | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H44
 
 | | Blasnase-P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H45
 
 | | Blasnase-T13A/P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H48
 
 | | Blasnase-T13A/P55F with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H49
 
 | | Blasnase-M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4A
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4C
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4G
 
 | | Blasnase-T13A/M57N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
5GWD
 
 | | Structure of Myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Xu, D, Ran, T, Wang, W. | | Deposit date: | 2016-09-10 | | Release date: | 2017-02-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
5GXV
 
 | | Crystal structure of PigG | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | Authors: | Zhang, F, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2016-09-20 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
7EW2
 
 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
