8K29
 
 | |
8K24
 
 | |
8K21
 
 | | Cas1-Cas2-dsDNA subregion in ICP1 Csy-DNA-Cas1-2/3 complex | | Descriptor: | Cas1, DNA (5'-D(*AP*TP*CP*TP*TP*CP*CP*CP*TP*AP*TP*TP*TP*AP*AP*AP*TP*TP*GP*CP*T)-3'), DNA (5'-D(P*AP*GP*CP*AP*AP*TP*TP*TP*AP*AP*AP*TP*AP*GP*GP*GP*AP*AP*GP*AP*T)-3'), ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of csy complex with short DNA
To Be Published
|
|
8K27
 
 | |
8K25
 
 | | Structure of Cas1-Cas2-dsDNA complex | | Descriptor: | Cas1, DNA (5'-D(P*CP*AP*AP*TP*TP*TP*AP*AP*AP*TP*AP*GP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*TP*TP*CP*CP*CP*TP*AP*TP*TP*TP*AP*AP*AP*TP*TP*GP*C)-3'), ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Cas1-Cas2 complex bound dsDNA
To Be Published
|
|
8K28
 
 | |
8K26
 
 | | Structure of Cas1-Cas2 complex | | Descriptor: | Cas1, HD Cas3-type domain-containing protein | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of csy complex with short DNA
To Be Published
|
|
7YDL
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
7YHH
 
 | |
7YHG
 
 | |
7YHF
 
 | |
7YHI
 
 | |
8JYU
 
 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Acyl-ACP Synthetase structure bound to Decanoyl-AMP
To Be Published
|
|
8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
7ECW
 
 | | The Csy-AcrIF14-dsDNA complex | | Descriptor: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ELM
 
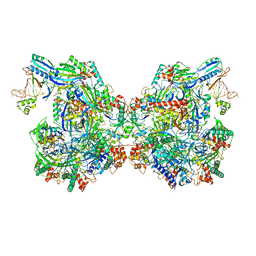 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
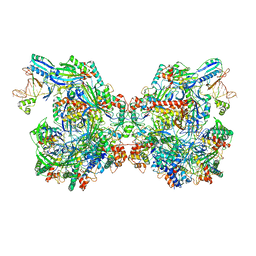 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7C78
 
 | |
8JD2
 
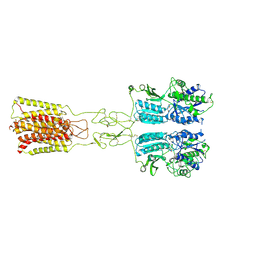 | | Cryo-EM structure of G protein-free mGlu2-mGlu3 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCW
 
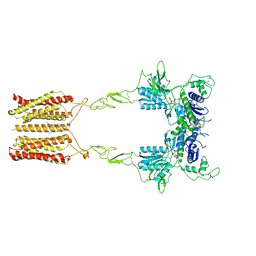 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD1
 
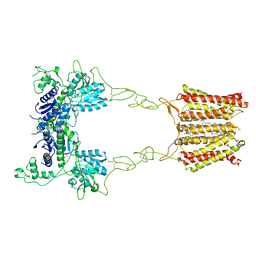 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in Rco state | | Descriptor: | CHOLESTEROL, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD6
 
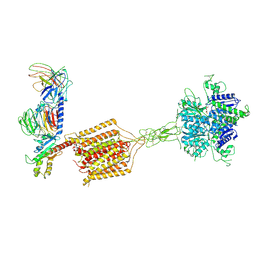 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCZ
 
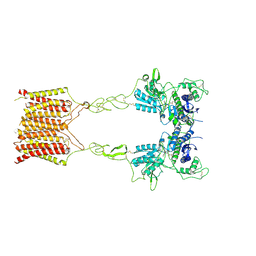 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCV
 
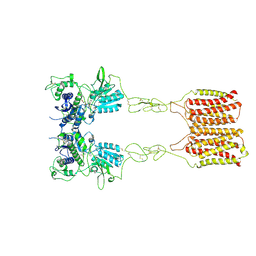 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
