8WLN
 
 | | Cryo-EM structure of the MS ring with export apparatus and proximal rod within the motor-hook complex in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-30 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKK
 
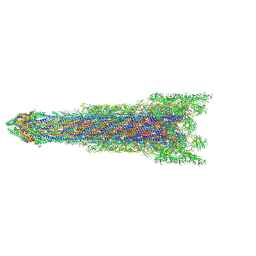 | | Cryo-EM structure of the whole rod with export apparatus and hook within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WKI
 
 | | Cryo-EM structure of the distal rod-hook within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-27 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
6LP7
 
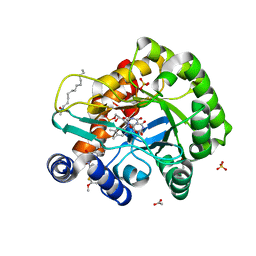 | | Crystal structure of human DHODH in complex with inhibitor 0944 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-(3-methoxyphenyl)phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2020-01-09 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
6LP6
 
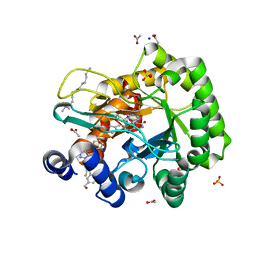 | | Crystal structure of human DHODH in complex with inhibitor 1214 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-[2-fluoranyl-5-(hydroxymethyl)phenyl]phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2020-01-09 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
9IJN
 
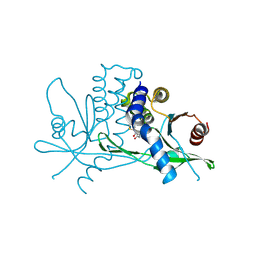 | | STING LBD domain with an agonist DW18343 | | Descriptor: | 4-[4-(7-fluoranyl-1H-indol-3-yl)thiophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Wang, Z. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a non-nucleotide stimulator of interferon genes (STING) agonist with systemic antitumor effect.
MedComm (2020), 6, 2025
|
|
8SX3
 
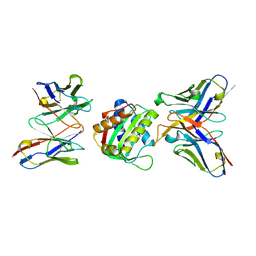 | | 10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10 | | Descriptor: | 10E8 Fab heavy chain, 10E8 light chain, 10E8-GT10.2 immunogen, ... | | Authors: | Huang, J, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Vaccination induces broadly neutralizing antibody precursors to HIV gp41.
Nat.Immunol., 25, 2024
|
|
8WZQ
 
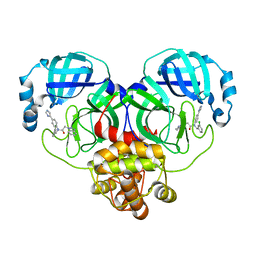 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Jiang, H.H, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WZP
 
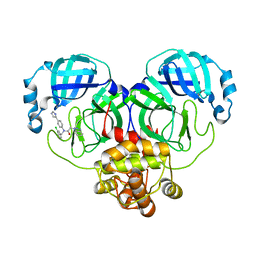 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
8WUL
 
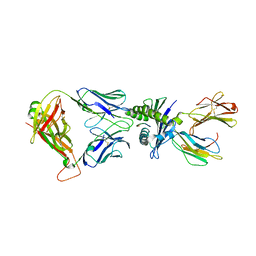 | | Crystal structure of affinity enhanced TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen, ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
8WTE
 
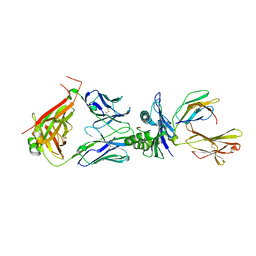 | | Crystal structure of TCR in complex with HLA-A*11:01 bound to KRAS-G12V peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12V nonamer peptide, MHC class I antigen (Fragment), ... | | Authors: | Zhang, M.Y, Luo, L.J, Xu, W, Guan, F.H, Wang, X.Y, Zhu, P, Zhang, J.H, Zhou, X.Y, Wang, F, Ye, S. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and affinity enhancement of T-cell receptor targeting a KRAS G12V cancer neoantigen.
Commun Biol, 7, 2024
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
5GNU
 
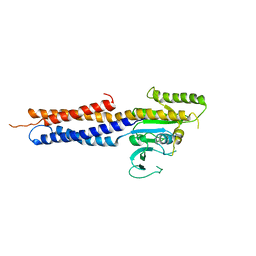 | | the structure of mini-MFN1 apo | | Descriptor: | Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
8HVX
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
6LTK
 
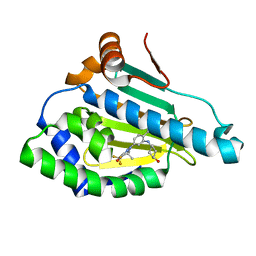 | | HSP90 in complex with SNX-2112 | | Descriptor: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
4E0R
 
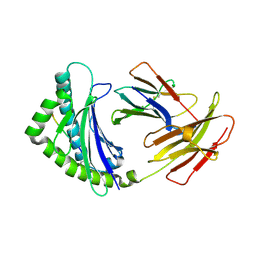 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
9IKQ
 
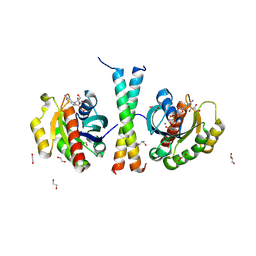 | |
9L0D
 
 | | Cryo-EM structure of the human MON1A/CCZ1/C18orf8 complex | | Descriptor: | Ras-related protein Rab-7a, Regulator of MON1-CCZ1 complex, Vacuolar fusion protein CCZ1 homolog B, ... | | Authors: | Tang, Y, Han, Y, Zhang, Y, Pan, L. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanistic insights into the GEF activity of the human MON1A/CCZ1/C18orf8 complex.
Protein Cell, 2025
|
|
4G42
 
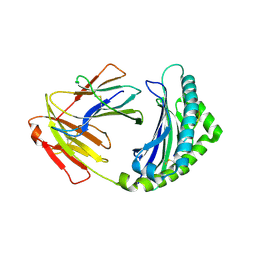 | | Structure of the Chicken MHC Class I Molecule BF2*0401 complexed to pepitde P8D | | Descriptor: | 8-MERIC PEPTIDE P8D, Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
5ZZ3
 
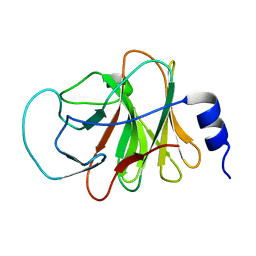 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | Descriptor: | Butyrophilin, subfamily 3, member A3 isoform b variant | | Authors: | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5IPC
 
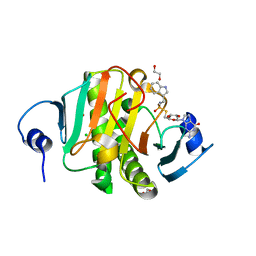 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
