5B6T
 
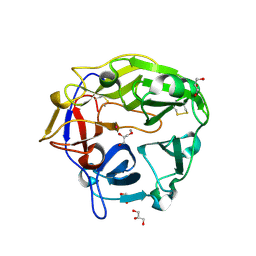 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase complexed with Pb | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
1WVO
 
 | | Solution structure of RSGI RUH-029, an antifreeze protein like domain in human N-acetylneuraminic acid phosphate synthase gene. | | Descriptor: | Sialic acid synthase | | Authors: | Ito, Y, Hamada, T, Hayashi, F, Yokoyama, S, Hirota, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-22 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the antifreeze-like domain of human sialic acid synthase
Protein Sci., 15, 2006
|
|
1RCH
 
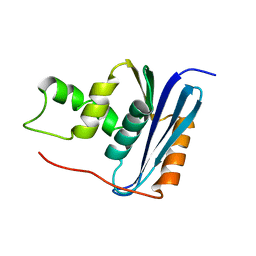 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
3ANR
 
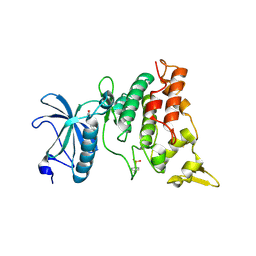 | | human DYRK1A/harmine complex | | Descriptor: | 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
3ANQ
 
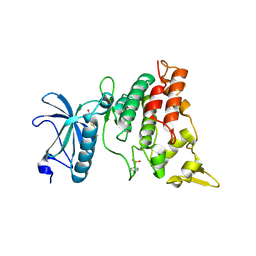 | | human DYRK1A/inhibitor complex | | Descriptor: | (1Z)-1-(3-ethyl-5-hydroxy-1,3-benzothiazol-2(3H)-ylidene)propan-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Nonaka, Y, Hosoya, T, Hagiwara, M, Ito, N. | | Deposit date: | 2010-09-06 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A
Nat Commun, 1, 2010
|
|
3AQA
 
 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2DCQ
 
 | |
2DCP
 
 | |
1VEE
 
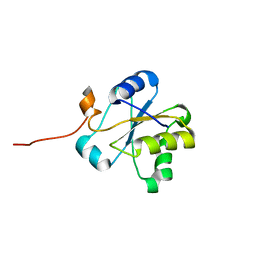 | | NMR structure of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana | | Descriptor: | proline-rich protein family | | Authors: | Pantoja-Uceda, D, Lopez-Mendez, B, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana
Protein Sci., 14, 2005
|
|
1OSJ
 
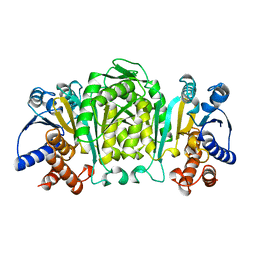 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
2D9D
 
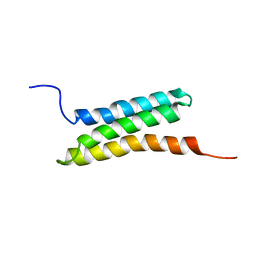 | | Solution structure of the BAG domain (275-350) of BAG-family molecular chaperone regulator-5 | | Descriptor: | BAG family molecular chaperone regulator 5 | | Authors: | Hatta, R, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1IPD
 
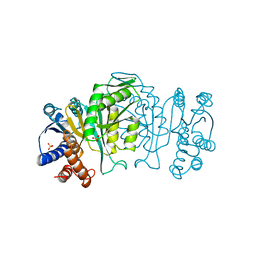 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
1OSI
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
8YD9
 
 | | Crystal structure of p38alpha with an allosteric inhibitor 3 | | Descriptor: | 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2024-02-19 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification of a novel target site for ATP-independent p38 alpha inhibitors
To Be Published
|
|
2E40
 
 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
2E3Z
 
 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in substrate-free form | | Descriptor: | Beta-glucosidase | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
1UK5
 
 | | Solution structure of the Murine BAG domain of Bcl2-associated athanogene 3 | | Descriptor: | BAG-family molecular chaperone regulator-3 | | Authors: | Hatta, R, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-19 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
1XAA
 
 | |
1XAB
 
 | |
3AKG
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKH
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKI
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKF
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
