7VXB
 
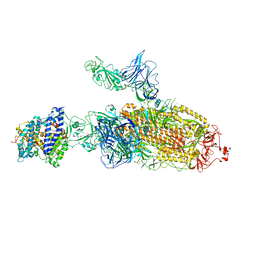 | | SARS-CoV-2 Kappa variant spike protein in C2b state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXK
 
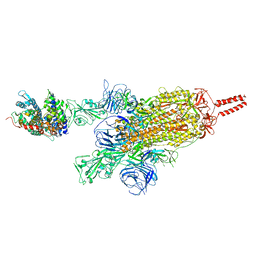 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2A state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXM
 
 | |
7WEV
 
 | |
7D9Q
 
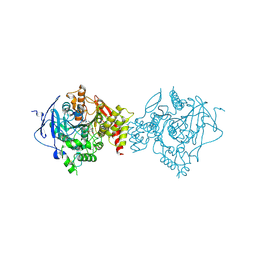 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 7 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(3-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9P
 
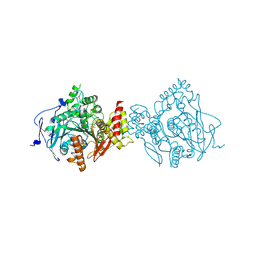 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 12 | | Descriptor: | (2S)-2-[[4-fluoranyl-1-[(2-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7D9O
 
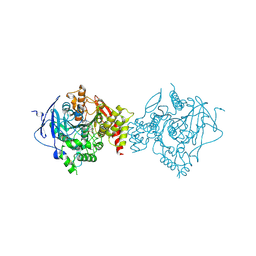 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
2LMZ
 
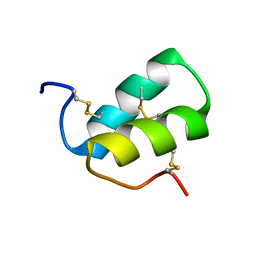 | |
7WK2
 
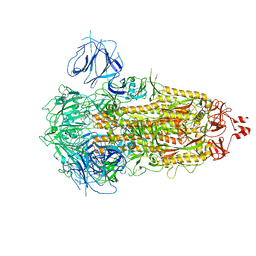 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
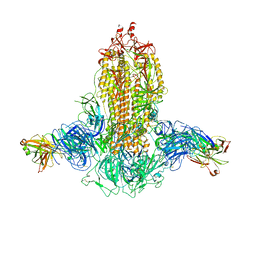
7WKA
 
 | |
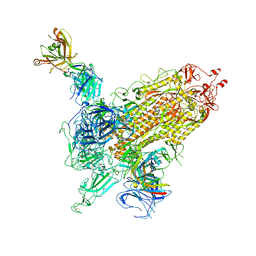
7WK9
 
 | |
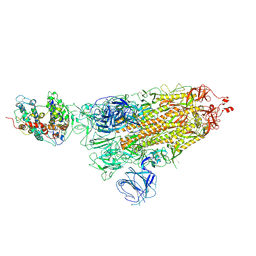
7WK4
 
 | |
7WK8
 
 | |
7WK6
 
 | |
7WVO
 
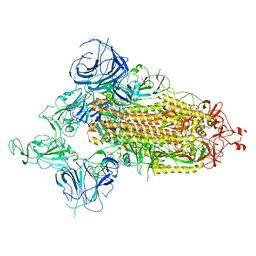 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
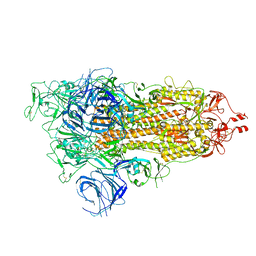 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
8G46
 
 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | Authors: | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
7DO1
 
 | |
7CYV
 
 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6J6F
 
 | | Ligand binding domain 1 and 2 of Talaromyces marneffei Mp1 protein | | Descriptor: | Envelope glycoprotein, NICKEL (II) ION | | Authors: | Lam, W.H, Zhang, H, Hao, Q. | | Deposit date: | 2019-01-15 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Talaromyces marneffeiMp1 Protein, a Novel Virulence Factor, Carries Two Arachidonic Acid-Binding Domains To Suppress Inflammatory Responses in Hosts.
Infect. Immun., 87, 2019
|
|
6IFE
 
 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
5XBW
 
 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5XBI
 
 | |
5XBT
 
 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
