8FBX
 
 | | Selenosugar synthase SenB from Variovorax paradoxus | | Descriptor: | CHLORIDE ION, SODIUM ION, Selenosugar synthase SenB | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Characterization and Ligand-Induced Conformational Changes of SenB, a Se-Glycosyltransferase Involved in Selenoneine Biosynthesis.
Biochemistry, 62, 2023
|
|
8FRZ
 
 | |
8FRW
 
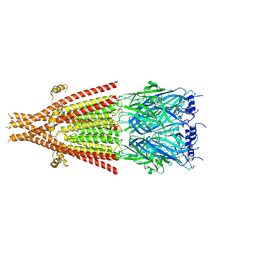 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FRX
 
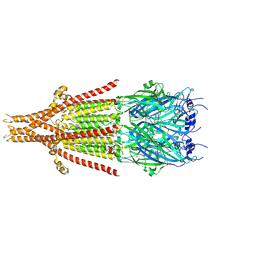 | | Full-length mouse 5-HT3A receptor in complex with SMP100, pre-activated | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FSZ
 
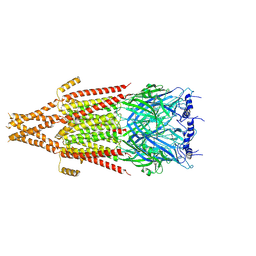 | | Full-length mouse 5-HT3A receptor in complex with ALB148471, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,5R)-1-azabicyclo[3.2.2]nonan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FSB
 
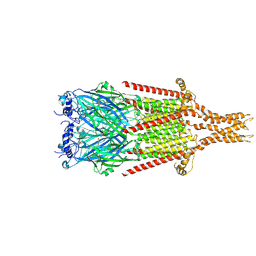 | |
8FSP
 
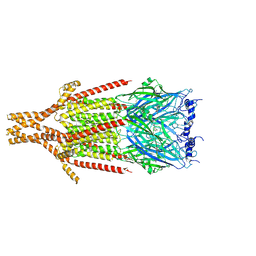 | | Full-length mouse 5-HT3A receptor in complex with SMP100, open-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl]-1,3,4,5-tetrahydro-6H-azepino[5,4,3-cd]indazol-6-one, 5-hydroxytryptamine receptor 3A | | Authors: | Felt, K.C, Chakrapani, S. | | Deposit date: | 2023-01-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for partial agonism in 5-HT 3A receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6LKC
 
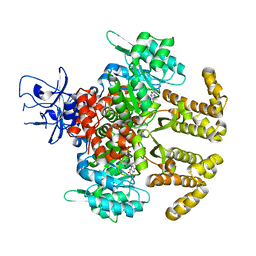 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
6LID
 
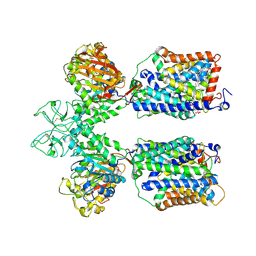 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
8IJS
 
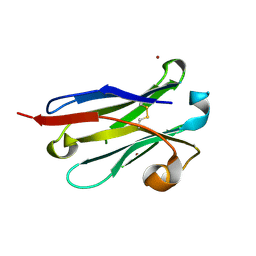 | | anti-VEGF nanobody mutant | | Descriptor: | ZINC ION, anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IIU
 
 | | anti-VEGF nanobody | | Descriptor: | SULFATE ION, anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IJZ
 
 | | anti-VEGF mutant | | Descriptor: | anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
6LI9
 
 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
5WPI
 
 | |
4OWT
 
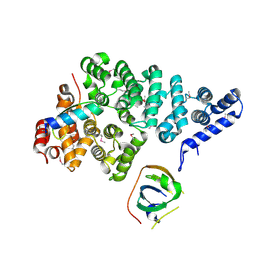 | | Structural basis of SOSS1 complex assembly | | Descriptor: | Integrator complex subunit 3, SOSS complex subunit B1, SOSS complex subunit C | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
4OWX
 
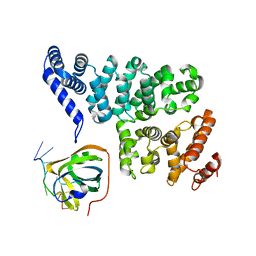 | | Structural basis of SOSS1 in complex with a 12nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1 | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
6IIC
 
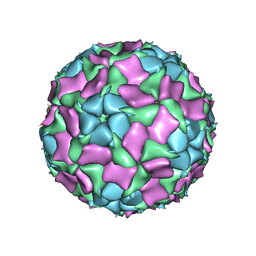 | | CryoEM structure of Mud Crab Dicistrovirus | | Descriptor: | VP1 of Mud crab dicistrovirus, VP2 of Mud crab dicistrovirus, VP3 of Mud crab dicistrovirus, ... | | Authors: | Zhang, Q, Gao, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Novel Viruses from Mud CrabScylla paramamosainwith Multiple Infections.
J. Virol., 93, 2019
|
|
5JC9
 
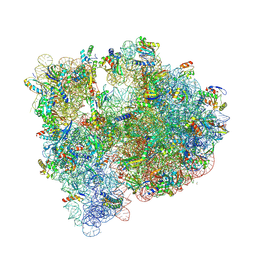 | | Structure of the Escherichia coli ribosome with the U1052G mutation in the 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-14 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7L
 
 | | Structure of the 70S E coli ribosome with the U1052G mutation in the 16S rRNA bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-07-27 | | Last modified: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JG8
 
 | | Crystal structure of human acid sphingomyelinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, Z.J, Prive, G.G. | | Deposit date: | 2016-04-19 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Human Acid Sphingomyelinase Reveals the Role of the Saposin Domain in Activating Substrate Hydrolysis.
J.Mol.Biol., 428, 2016
|
|
5J88
 
 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
