5I86
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
6MG7
 
 | | Crystal structure of the RV144 C1-C2 specific antibody CH54 Fab in complex with HIV-1 CLADE A/E GP120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH54 Fab heavy chain, CH54 Fab light chain, ... | | Authors: | Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6LQZ
 
 | | Solution structure of Taf14ET-Sth1EBMC | | Descriptor: | Nuclear protein STH1/NPS1, Transcription initiation factor TFIID subunit 14 | | Authors: | Wu, B, Chen, G, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Nat Commun, 11, 2020
|
|
2LDM
 
 | |
5I8G
 
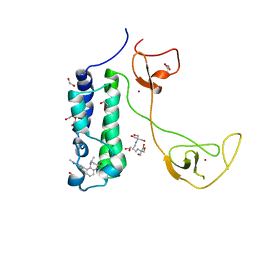 | | CBP in complex with Cpd637 ((R)-4-methyl-6-(1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-4-methyl-6-[1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I8B
 
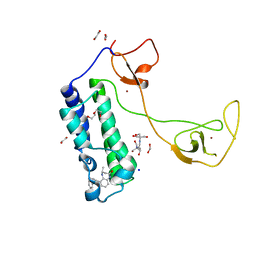 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
8J7N
 
 | |
8J7P
 
 | |
8HTD
 
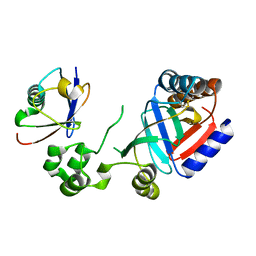 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTF
 
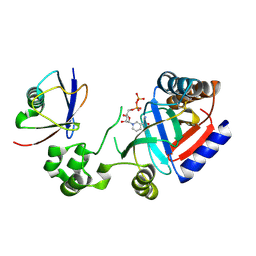 | | Crystal structure of an effector in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTC
 
 | | Crystal structure of a SeMet-labeled effector from Chromobacterium violaceum in complex with Ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin-40S ribosomal protein S27a (Fragment) | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
4M3O
 
 | | Crystal structure of K.lactis Rtr1 NTD | | Descriptor: | KLLA0F12672p, ZINC ION | | Authors: | Hsu, P.L, Yang, W, Zheng, N, Varani, G. | | Deposit date: | 2013-08-06 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rtr1 Is a Dual Specificity Phosphatase That Dephosphorylates Tyr1 and Ser5 on the RNA Polymerase II CTD.
J.Mol.Biol., 426, 2014
|
|
1CZJ
 
 | | CYTOCHROME C OF CLASS III (AMBLER) 26 KD | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Czjzek, M, Haser, R. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a dimeric octaheme cytochrome c3 (M(r) 26,000) from Desulfovibrio desulfuricans Norway.
Structure, 4, 1996
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
6KX2
 
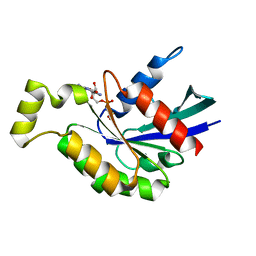 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
5JDK
 
 | |
8XQT
 
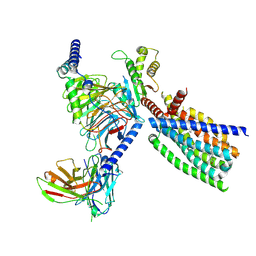 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQR
 
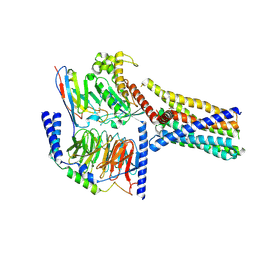 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQO
 
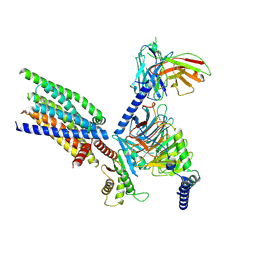 | | Structure of human class T GPCR TAS2R14-Gi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQS
 
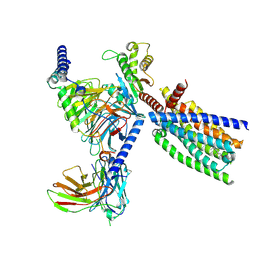 | | Structure of human class T GPCR TAS2R14-DNGi complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
3NMU
 
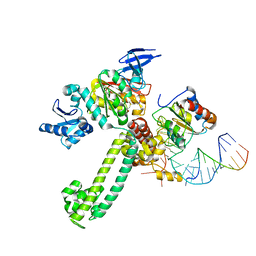 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NVK
 
 | | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Xue, S, Wang, R, Li, H. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NVM
 
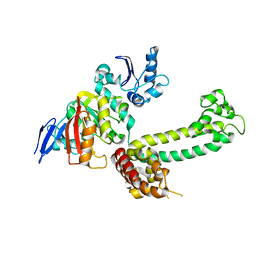 | |
